STEEP mediates STING ER exit and activation of signaling
- PMID: 32690950
- PMCID: PMC7610351
- DOI: 10.1038/s41590-020-0730-5
STEEP mediates STING ER exit and activation of signaling
Erratum in
-
Author Correction: STEEP mediates STING ER exit and activation of signaling.Nat Immunol. 2020 Nov;21(11):1468-1469. doi: 10.1038/s41590-020-0803-5. Nat Immunol. 2020. PMID: 32929276
Abstract
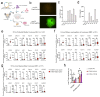
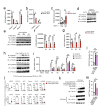
STING is essential for control of infections and for tumor immunosurveillance, but it can also drive pathological inflammation. STING resides on the endoplasmic reticulum (ER) and traffics following stimulation to the ERGIC/Golgi, where signaling occurs. Although STING ER exit is the rate-limiting step in STING signaling, the mechanism that drives this process is not understood. Here we identify STEEP as a positive regulator of STING signaling. STEEP was associated with STING and promoted trafficking from the ER. This was mediated through stimulation of phosphatidylinositol-3-phosphate (PtdIns(3)P) production and ER membrane curvature formation, thus inducing COPII-mediated ER-to-Golgi trafficking of STING. Depletion of STEEP impaired STING-driven gene expression in response to virus infection in brain tissue and in cells from patients with STING-associated diseases. Interestingly, STING gain-of-function mutants from patients interacted strongly with STEEP, leading to increased ER PtdIns(3)P levels and membrane curvature. Thus, STEEP enables STING signaling by promoting ER exit.
Conflict of interest statement
The authors declare no competing interests.
Figures














References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

