The activation trajectory of plasmacytoid dendritic cells in vivo during a viral infection
- PMID: 32690951
- PMCID: PMC7610367
- DOI: 10.1038/s41590-020-0731-4
The activation trajectory of plasmacytoid dendritic cells in vivo during a viral infection
Abstract
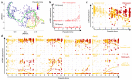
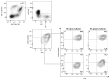
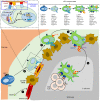
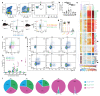
Plasmacytoid dendritic cells (pDCs) are a major source of type I interferon (IFN-I). What other functions pDCs exert in vivo during viral infections is controversial, and more studies are needed to understand their orchestration. In the present study, we characterize in depth and link pDC activation states in animals infected by mouse cytomegalovirus by combining Ifnb1 reporter mice with flow cytometry, single-cell RNA sequencing, confocal microscopy and a cognate CD4 T cell activation assay. We show that IFN-I production and T cell activation were performed by the same pDC, but these occurred sequentially in time and in different micro-anatomical locations. In addition, we show that pDC commitment to IFN-I production was marked early on by their downregulation of leukemia inhibitory factor receptor and was promoted by cell-intrinsic tumor necrosis factor signaling. We propose a new model for how individual pDCs are endowed to exert different functions in vivo during a viral infection, in a manner tightly orchestrated in time and space.
Conflict of interest statement
The authors declare no competing interests.
Figures












References
-
- Asselin-Paturel C, et al. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nature immunology. 2001;2:1144–1150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

