SPARC enables genetic manipulation of precise proportions of cells
- PMID: 32690967
- PMCID: PMC7939234
- DOI: 10.1038/s41593-020-0668-9
SPARC enables genetic manipulation of precise proportions of cells
Abstract
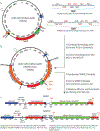
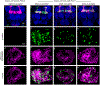
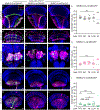
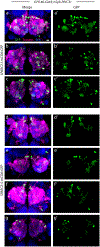
Many experimental approaches rely on controlling gene expression in select subsets of cells within an individual animal. However, reproducibly targeting transgene expression to specific fractions of a genetically defined cell type is challenging. We developed Sparse Predictive Activity through Recombinase Competition (SPARC), a generalizable toolkit that can express any effector in precise proportions of post-mitotic cells in Drosophila. Using this approach, we demonstrate targeted expression of many effectors in several cell types and apply these tools to calcium imaging of individual neurons and optogenetic manipulation of sparse cell populations in vivo.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

