Lysine acetylation regulates the RNA binding, subcellular localization and inclusion formation of FUS
- PMID: 32691043
- PMCID: PMC7530527
- DOI: 10.1093/hmg/ddaa159
Lysine acetylation regulates the RNA binding, subcellular localization and inclusion formation of FUS
Abstract
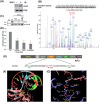
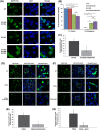
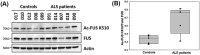
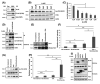
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by the preferential death of motor neurons. Approximately 10% of ALS cases are familial and 90% are sporadic. Fused in sarcoma (FUS) is a ubiquitously expressed RNA-binding protein implicated in familial ALS and frontotemporal dementia (FTD). The physiological function and pathological mechanism of FUS are not well understood, particularly whether post-translational modifications play a role in regulating FUS function. In this study, we discovered that FUS was acetylated at lysine-315/316 (K315/K316) and lysine-510 (K510) residues in two distinct domains. Located in the nuclear localization sequence, K510 acetylation disrupted the interaction between FUS and Transportin-1, resulting in the mislocalization of FUS in the cytoplasm and formation of stress granule-like inclusions. Located in the RNA recognition motif, K315/K316 acetylation reduced RNA binding to FUS and decreased the formation of cytoplasmic inclusions. Treatment with deacetylase inhibitors also significantly reduced the inclusion formation in cells expressing ALS mutation P525L. More interestingly, familial ALS patient fibroblasts showed higher levels of FUS K510 acetylation as compared with healthy controls. Lastly, CREB-binding protein/p300 acetylated FUS, whereas both sirtuins and histone deacetylases families of lysine deacetylases contributed to FUS deacetylation. These findings demonstrate that FUS acetylation regulates the RNA binding, subcellular localization and inclusion formation of FUS, implicating a potential role of acetylation in the pathophysiological process leading to FUS-mediated ALS/FTD.
Published by Oxford University Press 2020.
Figures


References
-
- Maruyama H., Morino H., Ito H., Izumi Y., Kato H., Watanabe Y., Kinoshita Y., Kamada M., Nodera H., Suzuki H. et al. (2010) Mutations of optineurin in amyotrophic lateral sclerosis. Nature, 465, 223–226. - PubMed
-
- Kwiatkowski T.J. Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T. et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science, 323, 1205–1208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

