Cyanobacterial metabolites as promising drug leads against the Mpro and PLpro of SARS-CoV-2: an in silico analysis
- PMID: 32691680
- PMCID: PMC7441779
- DOI: 10.1080/07391102.2020.1794972
Cyanobacterial metabolites as promising drug leads against the Mpro and PLpro of SARS-CoV-2: an in silico analysis
Abstract
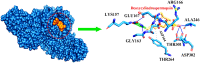
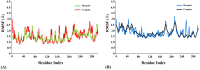
A novel severe acute respiratory syndrome coronavirus (SARS-CoV-2) has emerged as the causative agent behind the coronavirus disease 2019 (COVID-19) pandemic. Treatment efforts have been severely impeded due to the lack of specific effective antiviral drugs for the treatment of COVID-associated pathologies. In the present research endeavour the inhibitory prospects of cyanobacterial metabolites were assessed at the active binding pockets of the two vital SARS-CoV-2 proteases namely, main protease (Mpro) and the papain-like protease (PLpro) that proteolytically process viral polyproteins and facilitate viral replication, employing an in silico molecular interaction-based approach. It was evident from our analysis based on the binding energy scores that the metabolites cylindrospermopsin, deoxycylindrospermopsin, carrageenan, cryptophycin 52, eucapsitrione, tjipanazole, tolyporphin and apratoxin A exhibited promising inhibitory potential against the SARS-CoV-2 Mpro. The compounds cryptophycin 1, cryptophycin 52 and deoxycylindrospermopsin were observed to display encouraging binding energy scores with the PLpro of SARS-CoV-2. Subsequent estimation of physicochemical properties and potential toxicity of the metabolites followed by robust molecular dynamics simulations and analysis of MM-PBSA energy scoring function established deoxycylindrospermopsin as the most promising inhibitory candidate against both SARS-CoV-2 proteases. Present research findings bestow ample scopes to further exploit the potential of deoxycylindrospermopsin as a successful inhibitor of SARS-CoV-2 in vitro and in vivo and pave the foundation for the development of novel effective therapeutics against COVID-19.Communicated by Ramaswamy H. Sarma.
Keywords: MM-PBSA; SARS-CoV-2; cyanobacterial metabolites; deoxycylindrospermopsin; drug-likeness; molecular docking; molecular dynamics simulations.
Conflict of interest statement
The authors wish to declare that there are no conflicts of interest.
Figures



References
-
- Abdelli, I., Hassani, F., Bekkel Brikci, S., & Ghalem, S. (2020). In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1763199 - DOI - PMC - PubMed
-
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Adeoye, A. O., Oso, B. J., Olaoye, I. F., Tijjani, H., & Adebayo, A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1765876 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
