Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells
- PMID: 32691695
- PMCID: PMC7473213
- DOI: 10.1080/22221751.2020.1799723
Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells
Abstract
How severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infections engage cellular host pathways and innate immunity in infected cells remains largely elusive. We performed an integrative proteo-transcriptomics analysis in SARS-CoV-2 infected Huh7 cells to map the cellular response to the invading virus over time. We identified four pathways, ErbB, HIF-1, mTOR and TNF signaling, among others that were markedly modulated during the course of the SARS-CoV-2 infection in vitro. Western blot validation of the downstream effector molecules of these pathways revealed a dose-dependent activation of Akt, mTOR, S6K1 and 4E-BP1 at 24 hours post infection (hpi). However, we found a significant inhibition of HIF-1α through 24hpi and 48hpi of the infection, suggesting a crosstalk between the SARS-CoV-2 and the Akt/mTOR/HIF-1 signaling pathways. Inhibition of the mTOR signaling pathway using Akt inhibitor MK-2206 showed a significant reduction in virus production. Further investigations are required to better understand the molecular sequelae in order to guide potential therapy in the management of severe coronavirus disease 2019 (COVID-19) patients.
Keywords: Akt/mTOR/HIF-1; MK-2206; SARS-CoV-2; proteomics; transcriptomics.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



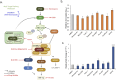
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
