Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19
- PMID: 32692185
- PMCID: PMC7640960
- DOI: 10.1021/acs.jproteome.0c00392
Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19
Abstract
SARS-CoV-2 is responsible for the current COVID-19 pandemic. On the basis of our analysis of hepatitis C virus and coronavirus replication, and the molecular structures and activities of viral inhibitors, we previously demonstrated that three nucleotide analogues (the triphosphates of Sofosbuvir, Alovudine, and AZT) inhibit the SARS-CoV RNA-dependent RNA polymerase (RdRp). We also demonstrated that a library of additional nucleotide analogues terminate RNA synthesis catalyzed by the SARS-CoV-2 RdRp, a well-established drug target for COVID-19. Here, we used polymerase extension experiments to demonstrate that the active triphosphate form of Sofosbuvir (an FDA-approved hepatitis C drug) is incorporated by SARS-CoV-2 RdRp and blocks further incorporation. Using the molecular insight gained from the previous studies, we selected the active triphosphate forms of six other antiviral agents, Alovudine, Tenofovir alafenamide, AZT, Abacavir, Lamivudine, and Emtricitabine, for evaluation as inhibitors of the SARS-CoV-2 RdRp and demonstrated the ability of these viral polymerase inhibitors to be incorporated by SARS-CoV-2 RdRp, where they terminate further polymerase extension with varying efficiency. These results provide a molecular basis for inhibition of the SARS-CoV-2 RdRp by these nucleotide analogues. If sufficient efficacy of some of these FDA-approved drugs in inhibiting viral replication in cell culture is established, they may be explored as potential COVID-19 therapeutics.
Keywords: COVID-19; RNA-dependent RNA polymerase; SARS-CoV-2; nucleotide analogues.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
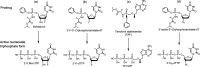
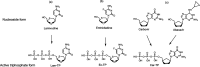
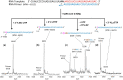


Update of
-
Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase.bioRxiv [Preprint]. 2020 Mar 20:2020.03.18.997585. doi: 10.1101/2020.03.18.997585. bioRxiv. 2020. Update in: J Proteome Res. 2020 Nov 6;19(11):4690-4697. doi: 10.1021/acs.jproteome.0c00392. PMID: 32511320 Free PMC article. Updated. Preprint.
References
-
- Zhu N.; Zhang D.; Wang W.; Li X.; Yang B.; Song J.; Zhao X.; Huang B.; Shi W.; Lu R.; Niu P.; Zhan F.; Ma X.; Wang D.; Xu W.; Wu G.; Gao G. F.; Tan W. for the China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Dustin L. B.; Bartolini B.; Capobianchi M. R.; Pistello M. Hepatitis C virus: life cycle in cells, infection and host response, and analysis of molecular markers influencing the outcome of infection and response to therapy. Clin. Microbiol. Infect. 2016, 22, 826–832. 10.1016/j.cmi.2016.08.025. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

