Single-Cell Reconstruction of Human Basal Cell Diversity in Normal and Idiopathic Pulmonary Fibrosis Lungs
- PMID: 32692579
- PMCID: PMC7706153
- DOI: 10.1164/rccm.201904-0792OC
Single-Cell Reconstruction of Human Basal Cell Diversity in Normal and Idiopathic Pulmonary Fibrosis Lungs
Abstract
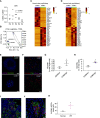
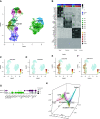
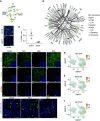
Rationale: Declining lung function in patients with interstitial lung disease is accompanied by epithelial remodeling and progressive scarring of the gas-exchange region. There is a need to better understand the contribution of basal cell hyperplasia and associated mucosecretory dysfunction to the development of idiopathic pulmonary fibrosis (IPF).Objectives: We sought to decipher the transcriptome of freshly isolated epithelial cells from normal and IPF lungs to discern disease-dependent changes within basal stem cells.Methods: Single-cell RNA sequencing was used to map epithelial cell types of the normal and IPF human airways. Organoid and air-liquid interface cultures were used to investigate functional properties of basal cell subtypes.Measurements and Main Results: We found that basal cells included multipotent and secretory primed subsets in control adult lung tissue. Secretory primed basal cells include an overlapping molecular signature with basal cells obtained from the distal lung tissue of IPF lungs. We confirmed that NOTCH2 maintains undifferentiated basal cells and restricts basal-to-ciliated differentiation, and we present evidence that NOTCH3 functions to restrain secretory differentiation.Conclusions: Basal cells are dynamically regulated in disease and are specifically biased toward the expansion of the secretory primed basal cell subset in IPF. Modulation of basal cell plasticity may represent a relevant target for therapeutic intervention in IPF.
Keywords: dynamic contrast enhanced magnetic resonance imaging; prematurity; pulmonary microvascular function; vascular simplification.
Figures


Comment in
-
Reproducible Single-Cell Genomic Research in Pulmonary and Critical Care Medicine.Am J Respir Crit Care Med. 2020 Dec 1;202(11):1495-1497. doi: 10.1164/rccm.202008-3073ED. Am J Respir Crit Care Med. 2020. PMID: 32916059 Free PMC article. No abstract available.
References
-
- Wells JM, Watt FM. Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature. 2018;557:322–328. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

