Bioinformatical dissection of fission yeast DNA replication origins
- PMID: 32692956
- PMCID: PMC7574548
- DOI: 10.1098/rsob.200052
Bioinformatical dissection of fission yeast DNA replication origins
Abstract
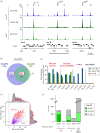
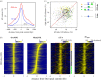
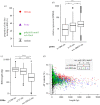
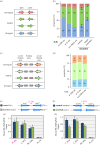
Replication origins in eukaryotes form a base for assembly of the pre-replication complex (pre-RC), thereby serving as an initiation site of DNA replication. Characteristics of replication origin vary among species. In fission yeast Schizosaccharomyces pombe, DNA of high AT content is a distinct feature of replication origins; however, it remains to be understood what the general molecular architecture of fission yeast origin is. Here, we performed ChIP-seq mapping of Orc4 and Mcm2, two representative components of the pre-RC, and described the characteristics of their binding sites. The analysis revealed that fission yeast efficient origins are associated with two similar but independent features: a ≥15 bp-long motif with stretches of As and an AT-rich region of a few hundred bp. The A-rich motif was correlated with chromosomal binding of Orc, a DNA-binding component in the pre-RC, whereas the AT-rich region was associated with efficient binding of the DNA replicative helicase Mcm. These two features, in combination with the third feature, a transcription-poor region of approximately 1 kb, enabled to distinguish efficient replication origins from the rest of chromosome arms with high accuracy. This study, hence, provides a model that describes how multiple functional elements specify DNA replication origins in fission yeast genome.
Keywords: ChIP-seq; fission yeast; machine learning; pre-replication complex; replication origins.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
