Molecular basis of ETV6-mediated predisposition to childhood acute lymphoblastic leukemia
- PMID: 32693409
- PMCID: PMC7819760
- DOI: 10.1182/blood.2020006164
Molecular basis of ETV6-mediated predisposition to childhood acute lymphoblastic leukemia
Abstract
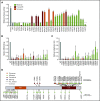
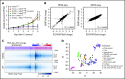
There is growing evidence supporting an inherited basis for susceptibility to acute lymphoblastic leukemia (ALL) in children. In particular, we and others reported recurrent germline ETV6 variants linked to ALL risk, which collectively represent a novel leukemia predisposition syndrome. To understand the influence of ETV6 variation on ALL pathogenesis, we comprehensively characterized a cohort of 32 childhood leukemia cases arising from this rare syndrome. Of 34 nonsynonymous germline ETV6 variants in ALL, we identified 22 variants with impaired transcription repressor activity, loss of DNA binding, and altered nuclear localization. Missense variants retained dimerization with wild-type ETV6 with potentially dominant-negative effects. Whole-transcriptome and whole-genome sequencing of this cohort of leukemia cases revealed a profound influence of germline ETV6 variants on leukemia transcriptional landscape, with distinct ALL subsets invoking unique patterns of somatic cooperating mutations. 70% of ALL cases with damaging germline ETV6 variants exhibited hyperdiploid karyotype with characteristic recurrent mutations in NRAS, KRAS, and PTPN11. In contrast, the remaining 30% cases had a diploid leukemia genome and an exceedingly high frequency of somatic copy-number loss of PAX5 and ETV6, with a gene expression pattern that strikingly mirrored that of ALL with somatic ETV6-RUNX1 fusion. Two ETV6 germline variants gave rise to both acute myeloid leukemia and ALL, with lineage-specific genetic lesions in the leukemia genomes. ETV6 variants compromise its tumor suppressor activity in vitro with specific molecular targets identified by assay for transposase-accessible chromatin sequencing profiling. ETV6-mediated ALL predisposition exemplifies the intricate interactions between inherited and acquired genomic variations in leukemia pathogenesis.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




Comment in
-
Germline ETV6 variants: not ALL created equally.Blood. 2021 Jan 21;137(3):288-289. doi: 10.1182/blood.2020008190. Blood. 2021. PMID: 33475741 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

