SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases
- PMID: 32694130
- PMCID: PMC7566278
- DOI: 10.1128/mSystems.00614-20
SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases
Abstract
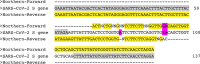
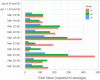
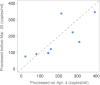
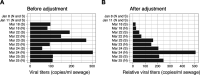
Wastewater surveillance represents a complementary approach to clinical surveillance to measure the presence and prevalence of emerging infectious diseases like the novel coronavirus SARS-CoV-2. This innovative data source can improve the precision of epidemiological modeling to understand the penetrance of SARS-CoV-2 in specific vulnerable communities. Here, we tested wastewater collected at a major urban treatment facility in Massachusetts and detected SARS-CoV-2 RNA from the N gene at significant titers (57 to 303 copies per ml of sewage) in the period from 18 to 25 March 2020 using RT-qPCR. We validated detection of SARS-CoV-2 by Sanger sequencing the PCR product from the S gene. Viral titers observed were significantly higher than expected based on clinically confirmed cases in Massachusetts as of 25 March. Our approach is scalable and may be useful in modeling the SARS-CoV-2 pandemic and future outbreaks.IMPORTANCE Wastewater-based surveillance is a promising approach for proactive outbreak monitoring. SARS-CoV-2 is shed in stool early in the clinical course and infects a large asymptomatic population, making it an ideal target for wastewater-based monitoring. In this study, we develop a laboratory protocol to quantify viral titers in raw sewage via qPCR analysis and validate results with sequencing analysis. Our results suggest that the number of positive cases estimated from wastewater viral titers is orders of magnitude greater than the number of confirmed clinical cases and therefore may significantly impact efforts to understand the case fatality rate and progression of disease. These data may help inform decisions surrounding the advancement or scale-back of social distancing and quarantine efforts based on dynamic wastewater catchment-level estimations of prevalence.
Keywords: COVID-19 prevalence; PMMoV; SARS-CoV-2; viral titers; wastewater.
Copyright © 2020 Wu et al.
Figures
Update of
-
SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases.medRxiv [Preprint]. 2020 Jul 6:2020.06.15.20117747. doi: 10.1101/2020.06.15.20117747. medRxiv. 2020. Update in: mSystems. 2020 Jul 21;5(4):e00614-20. doi: 10.1128/mSystems.00614-20. PMID: 32607521 Free PMC article. Updated. Preprint.
References
-
- Wang XW, Li J, Guo T, Zhen B, Kong Q, Yi B, Li Z, Song N, Jin M, Xiao W, Zhu X, Gu C, Yin J, Wei W, Yao W, Liu C, Li J, Ou G, Wang M, Fang T, Wang G, Qiu Y, Wu H, Chao F, Li J. 2005. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital of the Chinese People’s Liberation Army. Water Sci Technol 52:213–221. doi: 10.2166/wst.2005.0266. - DOI - PubMed
-
- Blomqvist S, Bassioni LE, Nasr EMEM, Paananen A, Kaijalainen S, Asghar H, de Gourville E, Roivainen M. 2012. Detection of imported wild polioviruses and of vaccine-derived polioviruses by environmental surveillance in Egypt. Appl Environ Microbiol 78:5406–5409. doi: 10.1128/AEM.00491-12. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

