A global lipid map defines a network essential for Zika virus replication
- PMID: 32694525
- PMCID: PMC7374707
- DOI: 10.1038/s41467-020-17433-9
A global lipid map defines a network essential for Zika virus replication
Abstract
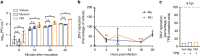
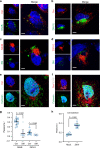
Zika virus (ZIKV), an arbovirus of global concern, remodels intracellular membranes to form replication sites. How ZIKV dysregulates lipid networks to allow this, and consequences for disease, is poorly understood. Here, we perform comprehensive lipidomics to create a lipid network map during ZIKV infection. We find that ZIKV significantly alters host lipid composition, with the most striking changes seen within subclasses of sphingolipids. Ectopic expression of ZIKV NS4B protein results in similar changes, demonstrating a role for NS4B in modulating sphingolipid pathways. Disruption of sphingolipid biosynthesis in various cell types, including human neural progenitor cells, blocks ZIKV infection. Additionally, the sphingolipid ceramide redistributes to ZIKV replication sites, and increasing ceramide levels by multiple pathways sensitizes cells to ZIKV infection. Thus, we identify a sphingolipid metabolic network with a critical role in ZIKV replication and show that ceramide flux is a key mediator of ZIKV infection.
Conflict of interest statement
The authors declare no competing interests.
Figures