Catecholamines suppress fatty acid re-esterification and increase oxidation in white adipocytes via STAT3
- PMID: 32694788
- PMCID: PMC7384260
- DOI: 10.1038/s42255-020-0217-6
Catecholamines suppress fatty acid re-esterification and increase oxidation in white adipocytes via STAT3
Abstract
Catecholamines stimulate the mobilization of stored triglycerides in adipocytes to provide fatty acids (FAs) for other tissues. However, a large proportion is taken back up and either oxidized or re-esterified. What controls the disposition of these FAs in adipocytes remains unknown. Here, we report that catecholamines redirect FAs for oxidation through the phosphorylation of signal transducer and activator of transcription 3 (STAT3). Adipocyte STAT3 is phosphorylated upon activation of β-adrenergic receptors, and in turn suppresses FA re-esterification to promote FA oxidation. Adipocyte-specific Stat3 KO mice exhibit normal rates of lipolysis, but exhibit defective lipolysis-driven oxidative metabolism, resulting in reduced energy expenditure and increased adiposity when they are on a high-fat diet. This previously unappreciated, non-genomic role of STAT3 explains how sympathetic activation can increase both lipolysis and FA oxidation in adipocytes, revealing a new regulatory axis in metabolism.
Conflict of interest statement
Competing Interests Statement:
The authors declare no competing interests.
Figures



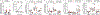
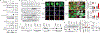



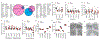
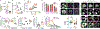
References
-
- Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ Tech Rep Ser 894, i–xii, 1–253 (2000). - PubMed
-
- Kennedy EP Biosynthesis of complex lipids. Fed Proc 20, 934–940 (1961). - PubMed
-
- Weiss SB, Kennedy EP & Kiyasu JY The enzymatic synthesis of triglycerides. J Biol Chem 235, 40–44 (1960). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

