Review of Viral Testing (Polymerase Chain Reaction) and Antibody/Serology Testing for Severe Acute Respiratory Syndrome-Coronavirus-2 for the Intensivist
- PMID: 32696013
- PMCID: PMC7314351
- DOI: 10.1097/CCE.0000000000000154
Review of Viral Testing (Polymerase Chain Reaction) and Antibody/Serology Testing for Severe Acute Respiratory Syndrome-Coronavirus-2 for the Intensivist
Abstract
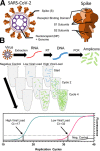
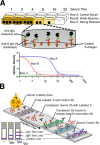
Objective: As the severe acute respiratory syndrome-coronavirus-2 pandemic develops, assays to detect the virus and infection caused by it are needed for diagnosis and management. To describe to clinicians how each assay is performed, what each assay detects, and the benefits and limitations of each assay.
Data sources: Published literature and internet.
Study selection: As well done, relevant and recent as possible.
Data extraction: Sources were read to extract data from them.
Data synthesis: Was synthesized by all coauthors.
Conclusions: Available assays test for current or previous severe acute respiratory syndrome-coronavirus-2 infection. Nucleic acid assays such as quantitative, or real-time, polymerase chain reaction and loop-mediated isothermal amplification are ideal for acute diagnosis with polymerase chain reaction testing remaining the "gold standard" to diagnose acute infection by severe acute respiratory syndrome-coronavirus-2, specifically the presence of viral RNA. Assays that detect serum antibodies can theoretically diagnose both acute and remote infection but require time for the patient to develop immunity and may detect nonspecific antibodies. Antibody assays that quantitatively measure neutralizing antibodies are needed to test efficacy of convalescent plasma therapy but are more specialized.
Keywords: coronavirus-2; molecular diagnostic techniques; neutralizing antibodies; real-time polymerase chain reaction; serologic tests; severe acute respiratory syndrome.
Copyright © 2020 The Authors. Published by Wolters Kluwer Health, Inc. on behalf of the Society of Critical Care Medicine.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

