Genetic and environmental determinants of variation in the plasma lipidome of older Australian twins
- PMID: 32697195
- PMCID: PMC7394543
- DOI: 10.7554/eLife.58954
Genetic and environmental determinants of variation in the plasma lipidome of older Australian twins
Abstract
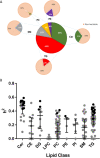
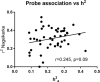
The critical role of blood lipids in a broad range of health and disease states is well recognised but less explored is the interplay of genetics and environment within the broader blood lipidome. We examined heritability of the plasma lipidome among healthy older-aged twins (75 monozygotic/55 dizygotic pairs) enrolled in the Older Australian Twins Study (OATS) and explored corresponding gene expression and DNA methylation associations. 27/209 lipids (13.3%) detected by liquid chromatography-coupled mass spectrometry (LC-MS) were significantly heritable under the classical ACE twin model (h2 = 0.28-0.59), which included ceramides (Cer) and triglycerides (TG). Relative to non-significantly heritable TGs, heritable TGs had a greater number of associations with gene transcripts, not directly associated with lipid metabolism, but with immune function, signalling and transcriptional regulation. Genome-wide average DNA methylation (GWAM) levels accounted for variability in some non-heritable lipids. We reveal a complex interplay of genetic and environmental influences on the ageing plasma lipidome.
Keywords: chromosomes; gene expression; genetics; genomics; heritability; human; lipidomics.
© 2020, Wong et al.
Conflict of interest statement
MW, AT, NB, KM, YL, LC, BB, NA, JK, PS, MW, DA, RP, TL, AP, PS No competing interests declared
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

