Khoe-San Genomes Reveal Unique Variation and Confirm the Deepest Population Divergence in Homo sapiens
- PMID: 32697301
- PMCID: PMC7530619
- DOI: 10.1093/molbev/msaa140
Khoe-San Genomes Reveal Unique Variation and Confirm the Deepest Population Divergence in Homo sapiens
Abstract
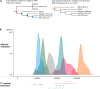
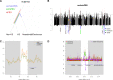
The southern African indigenous Khoe-San populations harbor the most divergent lineages of all living peoples. Exploring their genomes is key to understanding deep human history. We sequenced 25 full genomes from five Khoe-San populations, revealing many novel variants, that 25% of variants are unique to the Khoe-San, and that the Khoe-San group harbors the greatest level of diversity across the globe. In line with previous studies, we found several gene regions with extreme values in genome-wide scans for selection, potentially caused by natural selection in the lineage leading to Homo sapiens and more recent in time. These gene regions included immunity-, sperm-, brain-, diet-, and muscle-related genes. When accounting for recent admixture, all Khoe-San groups display genetic diversity approaching the levels in other African groups and a reduction in effective population size starting around 100,000 years ago. Hence, all human groups show a reduction in effective population size commencing around the time of the Out-of-Africa migrations, which coincides with changes in the paleoclimate records, changes that potentially impacted all humans at the time.
Keywords: Khoe-San; population structure; southern Africa.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Beuning KRM, Zimmerman KA, Ivory SJ, Cohen AS.. 2011. Vegetation response to glacial-interglacial climate variability near Lake Malawi in the southern African tropics. Palaeogeogr Palaeoclimatol Palaeoecol. 303(1–4):81–92.
-
- Breton G, Schlebusch CM, Lombard M, Sjodin P, Soodyall H, Jakobsson M.. 2014. Lactase persistence alleles reveal partial East African ancestry of southern African Khoe pastoralists. Curr Biol. 24(8):852–858. - PubMed
-
- Caley T, Extier T, Collins JA, Schefuß E, Dupont L, Malaizé B, Rossignol L, Souron A, McClymont EL, Jimenez-Espejo FJ, et al. 2018. A two-million-year-long hydroclimatic context for hominin evolution in southeastern Africa. Nature 560(7716):76–79. - PubMed
-
- Cann RL, Stoneking M, Wilson AC.. 1987. Mitochondrial DNA and human evolution. Nature 325(6099):31–36. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

