The Chemical Biology of Reversible Lysine Post-translational Modifications
- PMID: 32698016
- PMCID: PMC7487139
- DOI: 10.1016/j.chembiol.2020.07.002
The Chemical Biology of Reversible Lysine Post-translational Modifications
Abstract
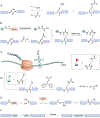
Lysine (Lys) residues in proteins undergo a wide range of reversible post-translational modifications (PTMs), which can regulate enzyme activities, chromatin structure, protein-protein interactions, protein stability, and cellular localization. Here we discuss the "writers," "erasers," and "readers" of some of the common protein Lys PTMs and summarize examples of their major biological impacts. We also review chemical biology approaches, from small-molecule probes to protein chemistry technologies, that have helped to delineate Lys PTM functions and show promise for a diverse set of biomedical applications.
Keywords: acetylation; acetyltransferase; bromodomain; deacetylase; enzyme; methylation; ubiquitination.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interests P.A.C. is a cofounder of Acylin and has been a scientific advisor for Abbvie, which has had therapeutics programs targeting p300 and CBP.
Figures


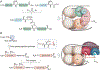

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

