Mid-Gestation lethality of Atxn2l-Ablated Mice
- PMID: 32698485
- PMCID: PMC7404131
- DOI: 10.3390/ijms21145124
Mid-Gestation lethality of Atxn2l-Ablated Mice
Abstract
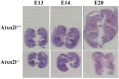
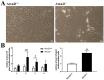
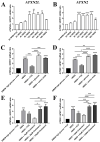
Depletion of yeast/fly Ataxin-2 rescues TDP-43 overexpression toxicity. In mouse models of Amyotrophic Lateral Sclerosis via TDP-43 overexpression, depletion of its ortholog ATXN2 mitigated motor neuron degeneration and extended lifespan from 25 days to >300 days. There is another ortholog in mammals, named ATXN2L (Ataxin-2-like), which is almost uncharacterized but also functions in RNA surveillance at stress granules. We generated mice with Crispr/Cas9-mediated deletion of Atxn2l exons 5-8, studying homozygotes prenatally and heterozygotes during aging. Our novel findings indicate that ATXN2L absence triggers mid-gestational embryonic lethality, affecting female animals more strongly. Weight and development stages of homozygous mutants were reduced. Placenta phenotypes were not apparent, but brain histology showed lamination defects and apoptosis. Aged heterozygotes showed no locomotor deficits or weight loss over 12 months. Null mutants in vivo displayed compensatory efforts to maximize Atxn2l expression, which were prevented upon nutrient abundance in vitro. Mouse embryonal fibroblast cells revealed more multinucleated giant cells upon ATXN2L deficiency. In addition, in human neural cells, transcript levels of ATXN2L were induced upon starvation and glucose and amino acids exposure, but this induction was partially prevented by serum or low cholesterol administration. Neither ATXN2L depletion triggered dysregulation of ATXN2, nor a converse effect was observed. Overall, this essential role of ATXN2L for embryogenesis raises questions about its role in neurodegenerative diseases and neuroprotective therapies.
Keywords: RNA chaperone; SCA2; Spinocerebellar ataxia type 2; fronto-temporal lobar dementia; nutrient endocytosis; poly(A)-tail; tauopathy; tyrosine kinase receptor signaling.
Conflict of interest statement
The authors declare no conflict of interest. G.A. advises RochePharma and TakedaPharma regarding ATXN2 research, receiving honoraria from them. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Fittschen M., Lastres-Becker I., Halbach M.V., Damrath E., Gispert S., Azizov M., Walter M., Muller S., Auburger G. Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics. 2015;16:181–192. doi: 10.1007/s10048-015-0441-5. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

