Emergence of European and North American mutant variants of SARS-CoV-2 in South-East Asia
- PMID: 32701194
- PMCID: PMC7405211
- DOI: 10.1111/tbed.13748
Emergence of European and North American mutant variants of SARS-CoV-2 in South-East Asia
Abstract
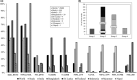
The SARS-CoV-2 coronavirus is responsible for the current COVID-19 pandemic, with an ongoing toll of over 5 million infections and 333 thousand deaths worldwide within the first 5 months. Insight into the phylodynamics and mutation variants of this virus is vital to understanding the nature of its spread in different climate conditions. The incidence rate of COVID-19 is increasing at an alarming pace within subtropical South-East Asian nations with high temperatures and humidity. To understand this spread, we analysed 444 genome sequences of SARS-CoV-2 available on the GISAID platform from six South-East Asian countries. Multiple sequence alignments and maximum-likelihood phylogenetic analyses were performed to analyse and characterize the non-synonymous (NS) mutant variants circulating in this region. Global mutation distribution analysis showed that the majority of the mutations found in this region are also prevalent in Europe and North America, and the concurrent presence of these mutations at a high frequency in other countries indicates possible transmission routes. Unique spike protein and non-structural protein mutations were observed circulating within confined area of a given country. We divided the circulating viral strains into four major groups and three subgroups on the basis of the most frequent NS mutations. Strains with a unique set of four co-evolving mutations were found to be circulating at a high frequency within India, specifically. Group 2 strains characterized by two co-evolving NS mutants which alter in RdRp (P323L) and spike (S) protein (D614G) were found to be common in Europe and North America. These European and North American variants have rapidly emerged as dominant strains within South-East Asia, increasing from a 0% prevalence in January to an 81% by May 2020. These variants may have an evolutionary advantage over their ancestral types and could present a large threat to South-East Asia for the coming winter.
Keywords: COVID-19; SARS-CoV-2; genome sequence; non-synonymous mutation; phylogeny.
© 2020 Wiley-VCH GmbH.
Conflict of interest statement
All authors declared that they do not have any conflicts of interests or relationship with this study financially or otherwise.
Figures




References
-
- Ayub, M. I. (2020). Reporting two SARS‐CoV‐2 strains based on a unique trinucleotide‐bloc mutation and their potential pathogenic difference. Preprints, 2020040337. Published online April, 19. 10.20944/preprints202004.0337.v1 - DOI
-
- Bajaj, P. , & Arya, P. C. (2020). Climatic‐niche evolution of SARS CoV‐2. bioRxiv. 10.1101/2020.06.18.147074 - DOI
-
- Bhattacharyya, C. , Das, C. , Ghosh, A. , Singh, A. K. , Mukherjee, S. , Majumder, P. P. , … Biswas, N. K. (2020). Global spread of SARS‐CoV‐2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. bioRxiv. 10.1101/2020.05.04.075911 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

