Transcriptomic Classification of Neurons Innervating Teeth
- PMID: 32702253
- PMCID: PMC7684839
- DOI: 10.1177/0022034520941837
Transcriptomic Classification of Neurons Innervating Teeth
Abstract
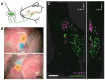
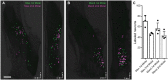
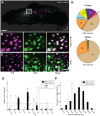
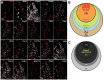
Toothache is a common painful consequence of damage to the teeth, particularly when coupled to infection. Clinical restoration of tooth integrity, sometimes involving physical and chemical sterilization of the tooth with nerve fiber ablation (i.e., endodontic therapy), generally alleviates pain and allows long-lasting dental function. These observations raise questions regarding the biological role of tooth-innervating fibers. Here, we determined the transcriptomic diversity of the sensory neurons that can be retrogradely labeled from mouse molar teeth. Our results demonstrate that individual molars are each targeted by a dedicated population of about 50 specialized trigeminal neurons. Transcriptomic profiling identifies the majority of these as expressing markers of fast-conducting neurons, with about two-thirds containing nociceptive markers. Our data provide a new view of dental innervation, extending previous reports that used candidate gene approaches. Importantly, almost all retrogradely labeled neurons, including nociceptors, express the recently characterized mechanosensor Piezo2, an ion channel that endows cells with sensitivity to gentle touch. Intriguingly, about a quarter of the labeled neurons do not appear to be nociceptors, perhaps insinuating a role for them in discriminative touch. We hypothesize that dental neurons are capable of providing mechanosensitive information to drive rapid behavioral responses and protect teeth from damage. Damage to the teeth and exposure of the large population of molar nociceptors may trigger prolonged or abnormal activation driving toothache. Future studies examining the responses of these transcriptomically defined classes of neurons will help define their significance in oral sensation.
Keywords: gene expression; mechanotransduction; neuroscience/neurobiology; pain; pulp biology; receptors.
Conflict of interest statement
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Figures

References
-
- Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP, Nassar MA, Dickenson AH, Wood JN. 2008. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 321(5889):702–705. - PubMed
-
- Chaudhary P, Martenson ME, Baumann TK. 2001. Vanilloid receptor expression and capsaicin excitation of rat dental primary afferent neurons. J Dent Res. 80(6):1518–1523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

