Microglia Require CD4 T Cells to Complete the Fetal-to-Adult Transition
- PMID: 32702313
- PMCID: PMC7427333
- DOI: 10.1016/j.cell.2020.06.026
Microglia Require CD4 T Cells to Complete the Fetal-to-Adult Transition
Abstract
The brain is a site of relative immune privilege. Although CD4 T cells have been reported in the central nervous system, their presence in the healthy brain remains controversial, and their function remains largely unknown. We used a combination of imaging, single cell, and surgical approaches to identify a CD69+ CD4 T cell population in both the mouse and human brain, distinct from circulating CD4 T cells. The brain-resident population was derived through in situ differentiation from activated circulatory cells and was shaped by self-antigen and the peripheral microbiome. Single-cell sequencing revealed that in the absence of murine CD4 T cells, resident microglia remained suspended between the fetal and adult states. This maturation defect resulted in excess immature neuronal synapses and behavioral abnormalities. These results illuminate a role for CD4 T cells in brain development and a potential interconnected dynamic between the evolution of the immunological and neurological systems. VIDEO ABSTRACT.
Keywords: CD4 T cells; T cells; brain; differentiation; human; microflora; microglia; migration; mouse; tissue-resident.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

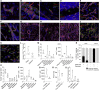




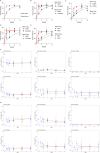

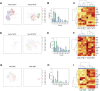




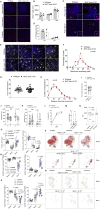

Comment in
-
T cells shape behaviour by helping microglia to mature.Nat Rev Immunol. 2020 Sep;20(9):518-519. doi: 10.1038/s41577-020-00411-z. Nat Rev Immunol. 2020. PMID: 32732953 No abstract available.
-
Microglia Get a Little Help from "Th"-eir Friends.Immunity. 2020 Sep 15;53(3):484-486. doi: 10.1016/j.immuni.2020.08.011. Immunity. 2020. PMID: 32937150
References
-
- Attaai A., Neidert N., von Ehr A., Potru P.S., Zöller T., Spittau B. Postnatal maturation of microglia is associated with alternative activation and activated TGFβ signaling. Glia. 2018;66:1695–1708. - PubMed
-
- Bechmann I., Galea I., Perry V.H. What is the blood-brain barrier (not)? Trends Immunol. 2007;28:5–11. - PubMed
-
- Belichenko P.V., Kleschevnikov A.M., Salehi A., Epstein C.J., Mobley W.C. Synaptic and cognitive abnormalities in mouse models of Down syndrome: exploring genotype-phenotype relationships. J. Comp. Neurol. 2007;504:329–345. - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

