TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells
- PMID: 32703818
- PMCID: PMC7383062
- DOI: 10.26508/lsa.202000786
TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells
Abstract
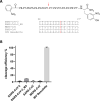
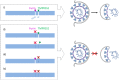
The novel emerged SARS-CoV-2 has rapidly spread around the world causing acute infection of the respiratory tract (COVID-19) that can result in severe disease and lethality. For SARS-CoV-2 to enter cells, its surface glycoprotein spike (S) must be cleaved at two different sites by host cell proteases, which therefore represent potential drug targets. In the present study, we show that S can be cleaved by the proprotein convertase furin at the S1/S2 site and the transmembrane serine protease 2 (TMPRSS2) at the S2' site. We demonstrate that TMPRSS2 is essential for activation of SARS-CoV-2 S in Calu-3 human airway epithelial cells through antisense-mediated knockdown of TMPRSS2 expression. Furthermore, SARS-CoV-2 replication was also strongly inhibited by the synthetic furin inhibitor MI-1851 in human airway cells. In contrast, inhibition of endosomal cathepsins by E64d did not affect virus replication. Combining various TMPRSS2 inhibitors with furin inhibitor MI-1851 produced more potent antiviral activity against SARS-CoV-2 than an equimolar amount of any single serine protease inhibitor. Therefore, this approach has considerable therapeutic potential for treatment of COVID-19.
© 2020 Bestle et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

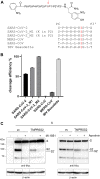
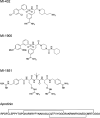
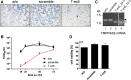

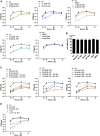
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
