Itaconate is an effector of a Rab GTPase cell-autonomous host defense pathway against Salmonella
- PMID: 32703879
- PMCID: PMC8020367
- DOI: 10.1126/science.aaz1333
Itaconate is an effector of a Rab GTPase cell-autonomous host defense pathway against Salmonella
Abstract
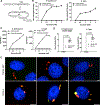
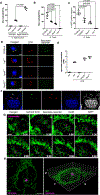
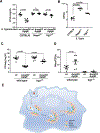
The guanosine triphosphatase (GTPase) Rab32 coordinates a cell-intrinsic host defense mechanism that restricts the replication of intravacuolar pathogens such as Salmonella Here, we show that this mechanism requires aconitate decarboxylase 1 (IRG1), which synthesizes itaconate, a metabolite with antimicrobial activity. We find that Rab32 interacts with IRG1 on Salmonella infection and facilitates the delivery of itaconate to the Salmonella-containing vacuole. Mice defective in IRG1 rescued the virulence defect of a S. enterica serovar Typhimurium mutant specifically defective in its ability to counter the Rab32 defense mechanism. These studies provide a link between a metabolite produced in the mitochondria after stimulation of innate immune receptors and a cell-autonomous defense mechanism that restricts the replication of an intracellular bacterial pathogen.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures
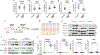
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

