Autophagy in kidney homeostasis and disease
- PMID: 32704047
- PMCID: PMC7868042
- DOI: 10.1038/s41581-020-0309-2
Autophagy in kidney homeostasis and disease
Abstract
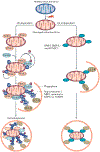
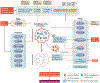
Autophagy is a conserved lysosomal pathway for the degradation of cytoplasmic components. Basal autophagy in kidney cells is essential for the maintenance of kidney homeostasis, structure and function. Under stress conditions, autophagy is altered as part of the adaptive response of kidney cells, in a process that is tightly regulated by signalling pathways that can modulate the cellular autophagic flux - mammalian target of rapamycin, AMP-activated protein kinase and sirtuins are key regulators of autophagy. Dysregulated autophagy contributes to the pathogenesis of acute kidney injury, to incomplete kidney repair after acute kidney injury and to chronic kidney disease of varied aetiologies, including diabetic kidney disease, focal segmental glomerulosclerosis and polycystic kidney disease. Autophagy also has a role in kidney ageing. However, questions remain about whether autophagy has a protective or a pathological role in kidney fibrosis, and about the precise mechanisms and signalling pathways underlying the autophagy response in different types of kidney cells and across the spectrum of kidney diseases. Further research is needed to gain insights into the regulation of autophagy in the kidneys and to enable the discovery of pathway-specific and kidney-selective therapies for kidney diseases and anti-ageing strategies.
Figures
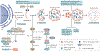


References
-
- Dikic I & Elazar Z Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol 19, 349–364 (2018). - PubMed
-
- Oku M & Sakai Y Three distinct types of microautophagy based on membrane dynamics and molecular machineries. Bioessays 40, e1800008 (2018). - PubMed
-
- Morishita H & Mizushima N Diverse cellular roles of autophagy. Annu. Rev. Cell Dev. Biol 35, 453–475 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

