Automated Sagittal Craniosynostosis Classification from CT Images Using Transfer Learning
- PMID: 32704611
- PMCID: PMC7377631
Automated Sagittal Craniosynostosis Classification from CT Images Using Transfer Learning
Abstract
Purpose: Sagittal Craniosynostosis (CSO) occurs when the sagittal suture of a growing child's skull is fused. Surgery is the primary treatment for CSO. Surgical treatment involves removing the affected bones and increasing the volume of the cranium by repositioning the bone segments or using external forces to guide growth. These external forces are often achieved by internal springs or external helmet therapy and depend on surgical judgment based on patient age, severity, and subtypes of CSO. Physicians usually classify CSO subtypes by examining CT images. In our previous work, we built an objective computerized system to mimic the physician's diagnostic process based on more than 100 hand-crafted features. However, hand-crafted features-based methods have limitations in representing all aspect features of the CSO images. To improve feature extraction efficiency, classification accuracy, and reduce subjectivity in the choice of surgical techniques, in this study, we developed a deep learning-based method to learn advanced features for the classification of CSO subtypes.
Methods: First, a Hounsfield Unit (HU) threshold-based method was used to segment 3D skulls from CT slices. Second, the 3D skulls were mapped to a two-dimension space by hemispherical projection to obtain binary images with a resolution of 512 × 512. These binary images were augmented to generate a new dataset for training deep convolutional neural networks. Finally, the pre-trained deep learning model was fine-tuned on the generated dataset using transfer learning method. Both training accuracy and cross-entropy curves were used to assess the performance of the proposed method.
Results: Three deep convolutional neural networks were built based on the manual classification results of CSO patients by three surgeons. The classification difference between surgeons was 54%. The prediction accuracy of the three deep learning models based on the generated dataset was greater than 90%, which was higher than the accuracy from the previous models (72%). The model based on the classification results of the senior surgeon achieved the highest performance accuracy (75%) in unseen real data, compared to 25% and 37.5% for two junior surgeons, respectively.
Conclusion: Our experimental results show that deep learning is superior to the hand-crafted feature-based method for sagittal CSO classification. The performance of deep learning models still depends on the quality of the original data. The classification variability of physicians can result in differential model outputs. When given more sagittal CSO images with proper annotations, the deep learning-based models can be more stable, approximate the diagnosis performance of physicians and have the potential to reduce the inter-observer variability thereby providing clinical insight into research and the treatment selection in patients with CSO.
Keywords: Convolutional neural networks; Medical image analysis; Sagittal craniosynostosis; Transfer learning.
Figures


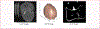





References
-
- Church MW, Parent-Jenkins L, Rozzelle AA, Eldis FE, Kazzi SNJ. Auditory brainstem response abnormalities and hearing loss in children with craniosynostosis. Pediatrics. 2007;119(6):e1351–60. - PubMed
-
- Kabbani H, Raghuveer TS. Craniosynostosis. American Family Physician. 2004;69(12):1–8. - PubMed
-
- Fellows-Mayle W, Hitchens TK, Simplaceanu E, Horner J, Barbano T, Losee JE, et al. Testing Causal mechanisms of nonsyn- dromic craniosynostosis using path analysis of cranial contents in rabbits with uncor- rected craniosynostosis. Cleft Palate-Craniofac J. 2006;43(5):524–31. - PubMed
-
- Kimonis V, Gold JA, Hoffman TL, Panchal J, Boyadjiev SA. Genetics of Craniosynostosis. In Seminars in Pediatric Neurology. Elsevier. 2007;14(3):150–61. - PubMed
-
- Ruiz-Correa S, Sze RW, Starr JR, Lin HTJ, Speltz ML, Cunningham ML, et al. New Scaphocephaly Severity Indices of Sagittal Craniosynostosis: A Comparative Study with Cranial Index Quantifications. Cleft Palate Craniofac J. 2006;43(2):211–21. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
