Comprehensive Genomic Analysis in NRG Oncology/RTOG 9802: A Phase III Trial of Radiation Versus Radiation Plus Procarbazine, Lomustine (CCNU), and Vincristine in High-Risk Low-Grade Glioma
- PMID: 32706640
- PMCID: PMC7527157
- DOI: 10.1200/JCO.19.02983
Comprehensive Genomic Analysis in NRG Oncology/RTOG 9802: A Phase III Trial of Radiation Versus Radiation Plus Procarbazine, Lomustine (CCNU), and Vincristine in High-Risk Low-Grade Glioma
Abstract
Purpose: NRG Oncology/RTOG 9802 (ClinicalTrials.gov Identifier: NCT00003375) is a practice-changing study for patients with WHO low-grade glioma (LGG, grade II), as it was the first to demonstrate a survival benefit of adjuvant chemoradiotherapy over radiotherapy. This post hoc study sought to determine the prognostic and predictive impact of the WHO-defined molecular subgroups and corresponding molecular alterations within NRG Oncology/RTOG 9802.
Methods: IDH1/2 mutations were determined by immunohistochemistry and/or deep sequencing. A custom Ion AmpliSeq panel was used for mutation analysis. 1p/19q codeletion and MGMT promoter methylation were determined by copy-number arrays and/or Illumina 450K array, respectively. Progression-free survival (PFS) and overall survival (OS) were estimated using the Kaplan-Meier method. Hazard ratios (HRs) were calculated using the Cox proportional hazard model and tested using the log-rank test. Multivariable analyses (MVAs) were performed incorporating treatment and common prognostic factors as covariates.
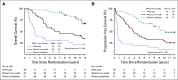
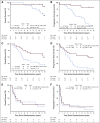
Results: Of the eligible patients successfully profiled for the WHO-defined molecular groups (n = 106/251), 26 (24%) were IDH-wild type, 43 (41%) were IDH-mutant/non-codeleted, and 37(35%) were IDH-mutant/codeleted. MVAs demonstrated that WHO subgroup was a significant predictor of PFS after adjustment for clinical variables and treatment. Notably, treatment with postradiation chemotherapy (PCV; procarbazine, lomustine (CCNU), and vincristine) was associated with longer PFS (HR, 0.32; P = .003; HR, 0.13; P < .001) and OS (HR, 0.38; P = .013; HR, 0.21; P = .029) in the IDH-mutant/non-codeleted and IDH-mutant/codeleted subgroups, respectively. In contrast, no significant difference in either PFS or OS was observed with the addition of PCV in the IDH-wild-type subgroup.
Conclusion: This study is the first to report the predictive value of the WHO-defined diagnostic classification in a set of uniformly treated patients with LGG in a clinical trial. Importantly, this post hoc analysis supports the notion that patients with IDH-mutant high-risk LGG regardless of codeletion status receive benefit from the addition of PCV.
Figures

Comment in
-
Back to the Future: Charting the Direction of Lower Grade Glioma Trials With Lessons From the Present and Past.Int J Radiat Oncol Biol Phys. 2022 Jan 1;112(1):30-34. doi: 10.1016/j.ijrobp.2021.10.002. Int J Radiat Oncol Biol Phys. 2022. PMID: 34919877 No abstract available.
References
-
- Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

