Increased Expression of Interleukin-1 Receptor Characterizes Anti-estrogen-Resistant ALDH+ Breast Cancer Stem Cells
- PMID: 32707076
- PMCID: PMC7419713
- DOI: 10.1016/j.stemcr.2020.06.020
Increased Expression of Interleukin-1 Receptor Characterizes Anti-estrogen-Resistant ALDH+ Breast Cancer Stem Cells
Abstract
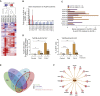
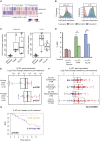
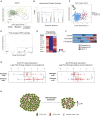
Estrogen-receptor-positive breast tumors are treated with anti-estrogen (AE) therapies but frequently develop resistance. Cancer stem cells (CSCs) with high aldehyde dehydrogenase activity (ALDH+ cells) are enriched following AE treatment. Here, we show that the interleukin-1β (IL-1β) signaling pathway is activated in ALDH+ cells, and data from single cells reveals that AE treatment selects for IL-1 receptor (IL1R1)-expressing ALDH+ cells. Importantly, CSC activity is reduced by an IL1R1 inhibitor in AE-resistant models. Moreover, IL1R1 expression is increased in the tumors of patients treated with AE therapy and predicts treatment failure. Single-cell gene expression analysis revealed that at least two subpopulations exist within the ALDH+ population, one proliferative and one quiescent. Following AE therapy the quiescent population is expanded, which suggests CSC dormancy as an adaptive strategy that facilitates treatment resistance. Targeting of ALDH+IL1R1+ cells merits testing as a strategy to combat AE resistance in patients with residual disease.
Keywords: ALDH(+) cells; IL1R1; anti-estrogens; breast cancer stem cells; dormancy.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Coser K.R., Wittner B.S., Rosenthal N.F., Collins S.C., Melas A., Smith S.L., Mahoney C.J., Shioda K., Isselbacher K.J., Ramaswamy S., Shioda T. Antiestrogen-resistant subclones of MCF-7 human breast cancer cells are derived from a common monoclonal drug-resistant progenitor. Proc. Natl. Acad. Sci. U S A. 2009;106:14536–14541. - PMC - PubMed
-
- Dunbier A.K., Ghazoui Z., Anderson H., Salter J., Nerurkar A., Osin P., A'Hern R., Miller W.R., Smith I.E., Dowsett M. Molecular profiling of aromatase inhibitor-treated postmenopausal breast tumors identifies immune-related correlates of resistance. Clin. Cancer Res. 2013;19:2775–2786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

