Fibroblast Growth Factor-14 Acts as Tumor Suppressor in Lung Adenocarcinomas
- PMID: 32707902
- PMCID: PMC7466013
- DOI: 10.3390/cells9081755
Fibroblast Growth Factor-14 Acts as Tumor Suppressor in Lung Adenocarcinomas
Abstract
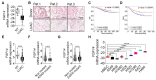
Investigation of the molecular dynamics in lung cancer is crucial for the development of new treatment strategies. Fibroblast growth factor (FGF) 14 belongs to the FGF family, which might play a crucial role in cancer progression. We analyzed lung adenocarcinoma (LUAC) patients samples and found that FGF14 was downregulated, correlating with reduced survival and oncogenic mutation status. FGF14 overexpression in lung cancer cell lines resulted in decreased proliferation, colony formation, and migration, as well as increased expression of epithelial markers and a decreased expression of mesenchymal markers, indicating a mesenchymal to epithelial transition in vitro. We verified these findings using small interfering RNA against FGF14 and further confirmed the suppressive effect of FGF14 in a NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ immunodeficient xenograft tumor model. Moreover, FGF14 overexpressing tumor cell RNA sequencing data suggests that genes affected by FGF14 were related to the extracellular matrix, playing a role in proliferation and migration. Notably, newly identified FGF14 target genes, adenosine deaminase RNA specific B1 (ADARB1), collagen and calcium-binding epidermal growth factor domain-containing protein 1 (CCBE1), α1 chain of collagen XI (COL11A1), and mucin 16 (MUC16) expression was negatively correlated with overall survival when FGF14 was downregulated in LUAC. These findings led us to suggest that FGF14 regulates proliferation and migration in LUAC.
Keywords: fibroblast growth factor 14; lung adenocarcinoma; lung cancer mesenchymal epithelial transition; xenograft model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Robert Koch-Institut; Gesellschaft der epidemiologischen Krebsregister in Deutschland e.V., editor. Krebs in Deutschland für 2015/2016 Krebs in Deutschland. Robert Koch-Institut; Gesellschaft der epidemiologischen Krebsregister in Deutschland e.V.; Berlin, Germany: 2017. 11. Ausgab.
-
- Aisner D., Sholl L.M., Berry L.D., Haura E.B., Ramalingam S.S., Glisson B.S., Socinski M.A., Waqar S.N., Garon E.B., Cetnar J.P., et al. Effect of expanded genomic testing in lung adenocarcinoma (LUCA) on survival benefit: The Lung Cancer Mutation Consortium II (LCMC II) experience. J. Clin. Oncol. 2016;34:11510. doi: 10.1200/JCO.2016.34.15_suppl.11510. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

