Leukaemia Inhibitory Factor (LIF) Inhibits Cancer Stem Cells Tumorigenic Properties through Hippo Kinases Activation in Gastric Cancer
- PMID: 32707998
- PMCID: PMC7464447
- DOI: 10.3390/cancers12082011
Leukaemia Inhibitory Factor (LIF) Inhibits Cancer Stem Cells Tumorigenic Properties through Hippo Kinases Activation in Gastric Cancer
Abstract
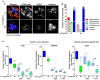
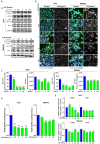
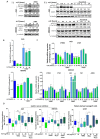
Cancer stem cells (CSCs) present chemo-resistance mechanisms contributing to tumour maintenance and recurrence, making their targeting of utmost importance in gastric cancer (GC) therapy. The Hippo pathway has been implicated in gastric CSC properties and was shown to be regulated by leukaemia inhibitory factor receptor (LIFR) and its ligand LIF in breast cancer. This study aimed to determine LIF's effect on CSC properties in GC cell lines and patient-derived xenograft (PDX) cells, which remains unexplored. LIF's treatment effect on CSC markers expression and tumoursphere formation was evaluated. The Hippo kinase inhibitor XMU-MP-1 and/or the JAK1 inhibitor Ruxolitinib were used to determine Hippo and canonical JAK/STAT pathway involvement in gastric CSCs' response to LIF. Results indicate that LIF decreased tumorigenic and chemo-resistant CSCs, in both GC cell lines and PDX cells. In addition, LIF increased activation of LATS1/2 Hippo kinases, thereby decreasing downstream YAP/TAZ nuclear accumulation and TEAD transcriptional activity. LIF's anti-CSC effect was reversed by XMU-MP-1 but not by Ruxolitinib treatment, highlighting the opposite effects of these two pathways downstream LIFR. In conclusion, LIF displays anti-CSC properties in GC, through Hippo kinases activation, and could in fine constitute a new CSCs-targeting strategy to help decrease relapse cases and bad prognosis in GC.
Keywords: ALDH; CD44; GP190; JAK; LATS1/2; Ruxolitinib; XMU-MP-1; YAP; gastric carcinoma.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Cancer Today. [(accessed on 9 March 2020)]; Available online: http://gco.iarc.fr/today/home.
-
- Knight W.R., Allum W.H. Gastric tumours. Medicine (Baltim.) 2019;47:309–313. doi: 10.1016/j.mpmed.2019.02.002. - DOI
-
- Nguyen P.H., Giraud J., Chambonnier L., Dubus P., Wittkop L., Belleannée G., Collet D., Soubeyran I., Evrard S., Rousseau B., et al. Characterization of Biomarkers of Tumorigenic and Chemoresistant Cancer Stem Cells in Human Gastric Carcinoma. Clin. Cancer Res. 2017;23:1586–1597. doi: 10.1158/1078-0432.CCR-15-2157. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

