hcapca: Automated Hierarchical Clustering and Principal Component Analysis of Large Metabolomic Datasets in R
- PMID: 32708222
- PMCID: PMC7407629
- DOI: 10.3390/metabo10070297
hcapca: Automated Hierarchical Clustering and Principal Component Analysis of Large Metabolomic Datasets in R
Abstract
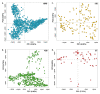
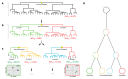
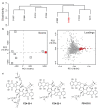
Microbial natural product discovery programs face two main challenges today: rapidly prioritizing strains for discovering new molecules and avoiding the rediscovery of already known molecules. Typically, these problems have been tackled using biological assays to identify promising strains and techniques that model variance in a dataset such as PCA to highlight novel chemistry. While these tools have shown successful outcomes in the past, datasets are becoming much larger and require a new approach. Since PCA models are dependent on the members of the group being modeled, large datasets with many members make it difficult to accurately model the variance in the data. Our tool, hcapca, first groups strains based on the similarity of their chemical composition, and then applies PCA to the smaller sub-groups yielding more robust PCA models. This allows for scalable chemical comparisons among hundreds of strains with thousands of molecular features. As a proof of concept, we applied our open-source tool to a dataset with 1046 LCMS profiles of marine invertebrate associated bacteria and discovered three new analogs of an established anticancer agent from one promising strain.
Keywords: HCA; LCMS; PCA; dendrogram; genomics; metabolites; open source; variance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

