Classification of Microarray Gene Expression Data Using an Infiltration Tactics Optimization (ITO) Algorithm
- PMID: 32708429
- PMCID: PMC7397166
- DOI: 10.3390/genes11070819
Classification of Microarray Gene Expression Data Using an Infiltration Tactics Optimization (ITO) Algorithm
Abstract
A number of different feature selection and classification techniques have been proposed in literature including parameter-free and parameter-based algorithms. The former are quick but may result in local maxima while the latter use dataset-specific parameter-tuning for higher accuracy. However, higher accuracy may not necessarily mean higher reliability of the model. Thus, generalized optimization is still a challenge open for further research. This paper presents a warzone inspired "infiltration tactics" based optimization algorithm (ITO)-not to be confused with the ITO algorithm based on the Itõ Process in the field of Stochastic calculus. The proposed ITO algorithm combines parameter-free and parameter-based classifiers to produce a high-accuracy-high-reliability (HAHR) binary classifier. The algorithm produces results in two phases: (i) Lightweight Infantry Group (LIG) converges quickly to find non-local maxima and produces comparable results (i.e., 70 to 88% accuracy) (ii) Followup Team (FT) uses advanced tuning to enhance the baseline performance (i.e., 75 to 99%). Every soldier of the ITO army is a base model with its own independently chosen Subset selection method, pre-processing, and validation methods and classifier. The successful soldiers are combined through heterogeneous ensembles for optimal results. The proposed approach addresses a data scarcity problem, is flexible to the choice of heterogeneous base classifiers, and is able to produce HAHR models comparable to the established MAQC-II results.
Keywords: cancer; classification; clustering; computational intelligence; ensembles; infiltration; infiltration tactics optimization algorithm; machine learning; microarray.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

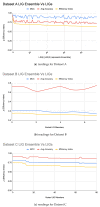
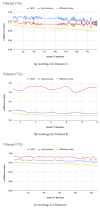
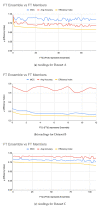
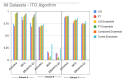
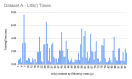
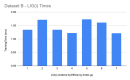
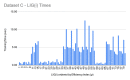
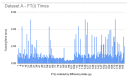
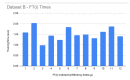
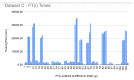
References
-
- Zhao Z., Morstatter F., Sharma S., Alelyani S., Anand A., Liu H. Advancing feature selection research. ASU Feature Sel. Repos. 2010:1–28.
-
- Elloumi M., Zomaya A.Y. Algorithms in Computational Molecular Biology: Techniques, Approaches and Applications. Volume 21 John Wiley & Sons; Hoboken, NJ, USA: 2011.
-
- Bolón-Canedo V., Sánchez-Marono N., Alonso-Betanzos A., Benítez J.M., Herrera F. A review of microarray datasets and applied feature selection methods. Inf. Sci. 2014;282:111–135. doi: 10.1016/j.ins.2014.05.042. - DOI
-
- Almugren N., Alshamlan H. A survey on hybrid feature selection methods in microarray gene expression data for cancer classification. IEEE Access. 2019;7:78533–78548. doi: 10.1109/ACCESS.2019.2922987. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

