miR-22-3p Negatively Affects Tumor Progression in T-Cell Acute Lymphoblastic Leukemia
- PMID: 32708470
- PMCID: PMC7408026
- DOI: 10.3390/cells9071726
miR-22-3p Negatively Affects Tumor Progression in T-Cell Acute Lymphoblastic Leukemia
Abstract
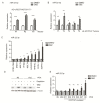
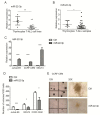
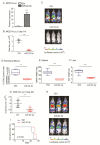
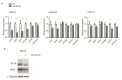
T-cell acute lymphoblastic leukemia (T-ALL) is a rare, aggressive disease arising from T-cell precursors. NOTCH1 plays an important role both in T-cell development and leukemia progression, and more than 60% of human T-ALLs harbor mutations in components of the NOTCH1 signaling pathway, leading to deregulated cell growth and contributing to cell transformation. Besides multiple NOTCH1 target genes, microRNAs have also been shown to regulate T-ALL initiation and progression. Using an established mouse model of T-ALL induced by NOTCH1 activation, we identified several microRNAs downstream of NOTCH1 activation. In particular, we found that NOTCH1 inhibition can induce miR-22-3p in NOTCH1-dependent tumors and that this regulation is also conserved in human samples. Importantly, miR-22-3p overexpression in T-ALL cells can inhibit colony formation in vitro and leukemia progression in vivo. In addition, miR-22-3p was found to be downregulated in T-ALL specimens, both T-ALL cell lines and primary samples, relative to immature T-cells. Our results suggest that miR-22-3p is a functionally relevant microRNA in T-ALL whose modulation can be exploited for therapeutic purposes to inhibit T-ALL progression.
Keywords: NOTCH1; T-ALL; miR-22-3p.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Gonzalez-Garcia S., Garcia-Peydro M., Martin-Gayo E., Ballestar E., Esteller M., Bornstein R., de la Pompa J.L., Ferrando A.A., Toribio M.L. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7R{alpha} gene expression in early human thymopoiesis and leukemia. J. Exp. Med. 2009;206:779–791. doi: 10.1084/jem.20081922. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

