Target-Genes Reveal Species and Genotypic Specificity of Anthocyanin Pigmentation in Citrus and Related Genera
- PMID: 32708660
- PMCID: PMC7397085
- DOI: 10.3390/genes11070807
Target-Genes Reveal Species and Genotypic Specificity of Anthocyanin Pigmentation in Citrus and Related Genera
Abstract
Background: Anthocyanin pigmentation characterizes a number of tissues of Citrus and its relatives. The gain and loss of pigmentation is intriguing and is inherited variously among species.
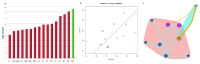
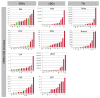
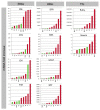
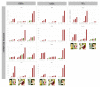
Methods: Citrus germplasm was used to investigate the anthocyanin pigmentation of tissues never before considered, including stamen, style and stigma, and of young leaves, petals, rind and flesh of 28 genotypes belonging to 14 species. Citrus genotypes encompassed citron, lemon, sweet orange, lime, and Citrus relatives included Microcitrus, Murraya, and Severinia. A relative qRT-PCR analysis was carried out on the structural and regulatory genes: phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3'-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), uridine diphosphate glucose flavonoid glucosyl-transferase (UFGT), glutathione S-transferase (GST), Ruby and Noemi. Image analysis and a genomic approach were employed to evaluate how the red pigmentation is inherited among tissues and species.
Results: Pigmentation of young leaves and petals is specific to citron and its hybrids. Ruby controls the pigmentation of petals, but not of leaves. The red color of the rind and flesh is a trait that particularly characterizes a diversity of sweet oranges, citron hybrids and Citrus relatives. Color expression depends on external factors and also on developmental stage. The coloration of stamen and style is citron-specific, while a red stigma is exclusive to Moro orange and its hybrids.
Conclusion: It is hypothesized that there is a relationship among Citrus species and genes controlling anthocyanin pigmentation.
Keywords: Citrus; fruit; image analysis; qRT-PCR; red color; stamen; stigma; style.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Ollitrault P., Curk F., Krueger R. Citrus Taxonomy. In: Talon M., Caruso M., Gmitter F.G., editors. The Genus Citrus. Woodhead Publishing, Elsevier; Duxford, UK: 2020. - DOI
-
- Li H., Deng Z., Zhu H., Hu C., Liu R., Young J.C., Tsao R. Highly Pigmented Vegetables: Anthocyanin Compositions and Their Role in Antioxidant Activities. Food Res. Int. 2012;46:250–259. doi: 10.1016/j.foodres.2011.12.014. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

