Brothers in Arms: Structure, Assembly and Function of Arenaviridae Nucleoprotein
- PMID: 32708976
- PMCID: PMC7411964
- DOI: 10.3390/v12070772
Brothers in Arms: Structure, Assembly and Function of Arenaviridae Nucleoprotein
Abstract
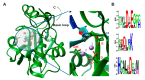
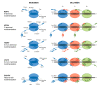
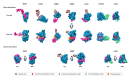
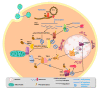
Arenaviridae is a family of viruses harbouring important emerging pathogens belonging to the Bunyavirales order. Like in other segmented negative strand RNA viruses, the nucleoprotein (NP) is a major actor of the viral life cycle being both (i) the necessary co-factor of the polymerase present in the L protein, and (ii) the last line of defence of the viral genome (vRNA) by physically hiding its presence in the cytoplasm. The NP is also one of the major players interfering with the immune system. Several structural studies of NP have shown that it features two domains: a globular RNA binding domain (NP-core) in its N-terminal and an exonuclease domain (ExoN) in its C-terminal. Further studies have observed that significant conformational changes are necessary for RNA encapsidation. In this review we revisited the most recent structural and functional data available on Arenaviridae NP, compared to other Bunyavirales nucleoproteins and explored the structural and functional implications. We review the variety of structural motif extensions involved in NP-NP binding mode. We also evaluate the major functional implications of NP interactome and the role of ExoN, thus making the NP a target of choice for future vaccine and antiviral therapy.
Keywords: Arenaviridae; Bunyavirales; emerging diseases; exonuclease; nucleoprotein; structure.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Ormay I., Kovács P. Lymphocytic choriomeningitis causing unilateral deafness. Orv. Hetil. 1989;130:789–791. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

