Inter-Regulation of Kv4.3 and Voltage-Gated Sodium Channels Underlies Predisposition to Cardiac and Neuronal Channelopathies
- PMID: 32709127
- PMCID: PMC7404392
- DOI: 10.3390/ijms21145057
Inter-Regulation of Kv4.3 and Voltage-Gated Sodium Channels Underlies Predisposition to Cardiac and Neuronal Channelopathies
Abstract
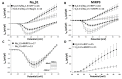
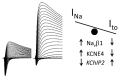
Background: Genetic variants in voltage-gated sodium channels (Nav) encoded by SCNXA genes, responsible for INa, and Kv4.3 channels encoded by KCND3, responsible for the transient outward current (Ito), contribute to the manifestation of both Brugada syndrome (BrS) and spinocerebellar ataxia (SCA19/22). We examined the hypothesis that Kv4.3 and Nav variants regulate each other's function, thus modulating INa/Ito balance in cardiomyocytes and INa/I(A) balance in neurons.
Methods: Bicistronic and other constructs were used to express WT or variant Nav1.5 and Kv4.3 channels in HEK293 cells. INa and Ito were recorded.
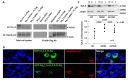
Results: SCN5A variants associated with BrS reduced INa, but increased Ito. Moreover, BrS and SCA19/22 KCND3 variants associated with a gain of function of Ito, significantly reduced INa, whereas the SCA19/22 KCND3 variants associated with a loss of function (LOF) of Ito significantly increased INa. Auxiliary subunits Navβ1, MiRP3 and KChIP2 also modulated INa/Ito balance. Co-immunoprecipitation and Duolink studies suggested that the two channels interact within the intracellular compartments and biotinylation showed that LOF SCN5A variants can increase Kv4.3 cell-surface expression.
Conclusion: Nav and Kv4.3 channels modulate each other's function via trafficking and gating mechanisms, which have important implications for improved understanding of these allelic cardiac and neuronal syndromes.
Keywords: Brugada syndrome; KCND3; Kv4.3; Nav1.1; Nav1.5; SCN1A; SCN5A; arrhythmia; channelopathies; spinocerebellar ataxia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Antzelevitch C., Yan G.-X., Ackerman M.J., Borggrefe M., Corrado D., Guo J., Gussak I., Hasdemir C., Horie M., Huikuri H., et al. J-Wave syndromes expert consensus conference report: Emerging concepts and gaps in knowledge. J. Arrhythmia. 2016;32:315–339. doi: 10.1016/j.joa.2016.07.002. - DOI - PMC - PubMed
-
- Clatot J., Ziyadeh-Isleem A., Maugenre S., Denjoy I., Liu H., Dilanian G., Hatem S.N., Deschênes I., Coulombe A., Guicheney P., et al. Dominant-negative effect of SCN5A N-terminal mutations through the interaction of Nav1.5 α-subunits. Cardiovasc. Res. 2012;96:53–63. doi: 10.1093/cvr/cvs211. - DOI - PMC - PubMed
-
- Giudicessi J.R., Ye D., Tester D.J., Crotti L., Mugione A., Nesterenko V.V., Albertson R.M., Antzelevitch C., Schwartz P.J., Ackerman M.J. Transient outward current (Ito) gain-of-function mutations in the KCND3-encoded Kv4.3 potassium channel and Brugada syndrome. Heart Rhythm. 2011;8:1024–1032. doi: 10.1016/j.hrthm.2011.02.021. - DOI - PMC - PubMed
-
- Duarri A., Nibbeling E., Fokkens M.R., Meijer M., Boddeke E., Lagrange E., Stevanin G., Brice A., Durr A., Verbeek D.S. The L450F [Corrected] mutation in KCND3 brings spinocerebellar ataxia and Brugada syndrome closer together. Neurogenetics. 2013;14:257–258. doi: 10.1007/s10048-013-0370-0. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

