Bifunctional Nitrone-Conjugated Secondary Metabolite Targeting the Ribosome
- PMID: 32709196
- PMCID: PMC8129991
- DOI: 10.1021/jacs.0c04675
Bifunctional Nitrone-Conjugated Secondary Metabolite Targeting the Ribosome
Abstract
Many microorganisms possess the capacity for producing multiple antibiotic secondary metabolites. In a few notable cases, combinations of secondary metabolites produced by the same organism are used in important combination therapies for treatment of drug-resistant bacterial infections. However, examples of conjoined roles of bioactive metabolites produced by the same organism remain uncommon. During our genetic functional analysis of oxidase-encoding genes in the everninomicin producer Micromonospora carbonacea var. aurantiaca, we discovered previously uncharacterized antibiotics everninomicin N and O, comprised of an everninomicin fragment conjugated to the macrolide rosamicin via a rare nitrone moiety. These metabolites were determined to be hydrolysis products of everninomicin P, a nitrone-linked conjugate likely the result of nonenzymatic condensation of the rosamicin aldehyde and the octasaccharide everninomicin F, possessing a hydroxylamino sugar moiety. Rosamicin binds the erythromycin macrolide binding site approximately 60 Å from the orthosomycin binding site of everninomicins. However, while individual ribosomal binding sites for each functional half of everninomicin P are too distant for bidentate binding, ligand displacement studies demonstrated that everninomicin P competes with rosamicin for ribosomal binding. Chemical protection studies and structural analysis of everninomicin P revealed that everninomicin P occupies both the macrolide- and orthosomycin-binding sites on the 70S ribosome. Moreover, resistance mutations within each binding site were overcome by the inhibition of the opposite functional antibiotic moiety binding site. These data together demonstrate a strategy for coupling orthogonal antibiotic pharmacophores, a surprising tolerance for substantial covalent modification of each antibiotic, and a potential beneficial strategy to combat antibiotic resistance.
Figures
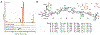

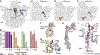


References
-
- Porse BT; Garrett RA, Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23S rRNA and the synergism of their inhibitory mechanisms. J Mol Biol 1999, 286 (2), 375–387. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- P60 DK020593/DK/NIDDK NIH HHS/United States
- P41 GM103391/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- S10 OD012359/OD/NIH HHS/United States
- T32 GM152284/GM/NIGMS NIH HHS/United States
- R01 GM092218/GM/NIGMS NIH HHS/United States
- R01 AI140400/AI/NIAID NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- T32 HL007751/HL/NHLBI NIH HHS/United States
- T32 GM007347/GM/NIGMS NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- F30 CA236131/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- T32 GM065086/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

