The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer
- PMID: 32709715
- PMCID: PMC7641983
- DOI: 10.1158/1078-0432.CCR-20-1825
The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer
Abstract
Purpose: SMARCA4 mutations are among the most common recurrent alterations in non-small cell lung cancer (NSCLC), but the relationship to other genomic abnormalities and clinical impact has not been established.
Experimental design: To characterize SMARCA4 alterations in NSCLC, we analyzed the genomic, protein expression, and clinical outcome data of patients with SMARCA4 alterations treated at Memorial Sloan Kettering.
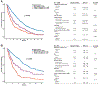
Results: In 4,813 tumors from patients with NSCLC, we identified 8% (n = 407) of patients with SMARCA4-mutant lung cancer. We describe two categories of SMARCA4 mutations: class 1 mutations (truncating mutations, fusions, and homozygous deletion) and class 2 mutations (missense mutations). Protein expression loss was associated with class 1 mutation (81% vs. 0%, P < 0.001). Both classes of mutation co-occurred more frequently with KRAS, STK11, and KEAP1 mutations compared with SMARCA4 wild-type tumors (P < 0.001). In patients with metastatic NSCLC, SMARCA4 alterations were associated with shorter overall survival, with class 1 alterations associated with shortest survival times (P < 0.001). Conversely, we found that treatment with immune checkpoint inhibitors (ICI) was associated with improved outcomes in patients with SMARCA4-mutant tumors (P = 0.01), with class 1 mutations having the best response to ICIs (P = 0.027).
Conclusions: SMARCA4 alterations can be divided into two clinically relevant genomic classes associated with differential protein expression as well as distinct prognostic and treatment implications. Both classes co-occur with KEAP1, STK11, and KRAS mutations, but individually represent independent predictors of poor prognosis. Despite association with poor outcomes, SMARCA4-mutant lung cancers may be more sensitive to immunotherapy.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures


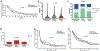
References
-
- Lovly CM, Gupta A, Lipson D, Otto G, Brennan T, Chung CT, et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov 2014;4(8):889–95 doi 10.1158/2159-8290.CD-14-0377. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

