Long-lasting severe immune dysfunction in Ebola virus disease survivors
- PMID: 32709840
- PMCID: PMC7381622
- DOI: 10.1038/s41467-020-17489-7
Long-lasting severe immune dysfunction in Ebola virus disease survivors
Abstract
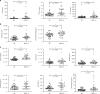
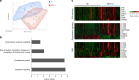
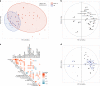
Long-term follow up studies from Ebola virus disease (EVD) survivors (EBOV_S) are lacking. Here, we evaluate immune and gene expression profiles in 35 Guinean EBOV_S from the last West African outbreak, a median of 23 months (IQR [18-25]) after discharge from treatment center. Compared with healthy donors, EBOV_S exhibit increases of blood markers of inflammation, intestinal tissue damage, T cell and B cell activation and a depletion of circulating dendritic cells. All survivors have EBOV-specific IgG antibodies and robust and polyfunctional EBOV-specific memory T-cell responses. Deep sequencing of the genes expressed in blood reveals an enrichment in 'inflammation' and 'antiviral' pathways. Integrated analyses identify specific immune markers associated with the persistence of clinical symptoms. This study identifies a set of biological and genetic markers that could be used to define a signature of "chronic Ebola virus disease (CEVD)".
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- WHO. Ebola Data and Statistics. Situation Summary.http://apps.who.int/gho/data/view.ebola-sitrep.ebola-summary-20160511?la... (2016).
-
- Baize S, et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014;371:1418–1425. - PubMed
-
- Etard J-F, et al. Multidisciplinary assessment of post-Ebola sequelae in Guinea (Postebogui): an observational cohort study. Lancet Infect. Dis. 2017;17:545–552. - PubMed
-
- Msellati P, et al. Revival after Ebola: multidisciplinary assessment at 1 year, prospect and follow-up study of surviving patients from Ebola in Guinea (PostEboGui cohort)] Bull. Soc. Pathol. Exot. 2016;109:236–243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

