Cyclin D1 targets hexokinase 2 to control aerobic glycolysis in myeloma cells
- PMID: 32709889
- PMCID: PMC7381668
- DOI: 10.1038/s41389-020-00253-3
Cyclin D1 targets hexokinase 2 to control aerobic glycolysis in myeloma cells
Abstract
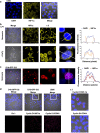
Cancer cells are characterized by the Warburg effect, a shift from mitochondrial respiration to oxidative glycolysis. We report here the crucial role of cyclin D1 in promoting this effect in a cyclin-dependent kinase (CDK)4/6-independent manner in multiple myeloma (MM) cells. We show that the cyclin D1 oncoprotein targets hexokinase 2 (HK2), a major glycolysis regulator, through two original molecular mechanisms in the cytoplasmic and nuclear compartments. In the cytoplasm, cyclin D1 binds HK2 at the outer mitochondrial membrane, and in the nucleus, it binds hypoxia-inducible factor-1α (HIF1α), which regulates HK2 gene transcription. We also show that high levels of HK2 expression are correlated with shorter event-free survival (EFS) and overall survival (OS) in MM patients. HK2 may therefore be considered as a possible target for antimyeloma therapy.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Ely S, et al. Mutually exclusive cyclin-dependent kinase 4/cyclin D1 and cyclin-dependent kinase 6/cyclin D2 pairing inactivates retinoblastoma protein and promotes cell cycle dysregulation in multiple myeloma. Clin. Cancer Res. 2005;65:11345–11353. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

