Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity
- PMID: 32711254
- PMCID: PMC7375792
- DOI: 10.1016/j.ebiom.2020.102911
Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity
Abstract
Background: Given the unceasing worldwide surge in COVID-19 cases, there is an imperative need to develop highly specific and sensitive serology assays to define exposure to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2).
Methods: Pooled plasma samples from PCR positive COVID-19 patients were used to identify linear B-cell epitopes from a SARS-CoV-2 peptide library of spike (S), envelope (E), membrane (M), and nucleocapsid (N) structural proteins by peptide-based ELISA. Hit epitopes were further validated with 79 COVID-19 patients with different disease severity status, 13 seasonal human CoV, 20 recovered SARS patients and 22 healthy donors.
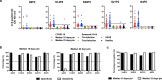
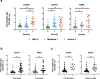
Findings: Four immunodominant epitopes, S14P5, S20P2, S21P2 and N4P5, were identified on the S and N viral proteins. IgG responses to all identified epitopes displayed a strong detection profile, with N4P5 achieving the highest level of specificity (100%) and sensitivity (>96%) against SARS-CoV-2. Furthermore, the magnitude of IgG responses to S14P5, S21P2 and N4P5 were strongly associated with disease severity.
Interpretation: IgG responses to the peptide epitopes can serve as useful indicators for the degree of immunopathology in COVID-19 patients, and function as higly specific and sensitive sero-immunosurveillance tools for recent or past SARS-CoV-2 infections. The flexibility of these epitopes to be used alone or in combination will allow for the development of improved point-of-care-tests (POCTs).
Funding: Biomedical Research Council (BMRC), the A*ccelerate GAP-funded project (ACCL/19-GAP064-R20H-H) from Agency of Science, Technology and Research (A*STAR), and National Medical Research Council (NMRC) COVID-19 Research fund (COVID19RF-001) and CCGSFPOR20002. ATR is supported by the Singapore International Graduate Award (SINGA), A*STAR.
Keywords: Biomarkers; COVID-19; Epitopes; Patients; SARS-CoV-2.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest SNA, CYPL, GC, CMP, LR and LFPN have filed a technology disclosure on the identified peptides with the patent application number 10202002981P. All other authors declare no conflicts.
Figures

Comment in
-
Immunodominant epitopes based serological assay for detecting SARS-CoV-2 exposure: Promises and challenges.EBioMedicine. 2020 Sep;59:102947. doi: 10.1016/j.ebiom.2020.102947. Epub 2020 Aug 15. EBioMedicine. 2020. PMID: 32807701 Free PMC article. No abstract available.
References
-
- Cohen J., Normile D. New SARS-like virus in China triggers alarm. Science. 2020;367(6475):234. - PubMed
-
- WHO. Coronavirus disease (COVID-2019) situation reports. 2020. Available from:https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio....
-
- Long Q-x, Deng H-j, Chen J., Hu J., Liu B-z, Liao P. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020;26(6):845–848. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

