HCV Replicon Systems: Workhorses of Drug Discovery and Resistance
- PMID: 32714881
- PMCID: PMC7344236
- DOI: 10.3389/fcimb.2020.00325
HCV Replicon Systems: Workhorses of Drug Discovery and Resistance
Abstract
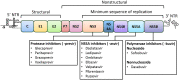
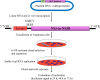
The development of direct-acting antivirals (DAAs) has revolutionized the state-of-the art treatment of HCV infections, with sustained virologic response rates above 90%. However, viral variants harboring substitutions referred to as resistance-associated substitutions (RASs) may be present in baseline levels and confer resistance to DAAs, thereby posing a major challenge for HCV treatment. HCV replicons have been the primary tools for discovering and evaluating the inhibitory activity of DAAs against viral replication. Interest in replicon systems has further grown as they have become indispensable for discovering genotype-specific and cross-genotype RASs. Here, we review functional replicon systems for HCV, how these replicon systems have contributed to the development of DAAs, and the characteristics and distribution of RASs for DAAs.
Keywords: direct-acting antiviral; drug discovery; drug resistance; genotype; hepatitis C virus; replicon; resistance-associated substitution.
Copyright © 2020 Khan, Soni and Veerapu.
Figures

Similar articles
-
Construction and characterization of Genotype-3 hepatitis C virus replicon revealed critical genotype-3-specific polymorphism for drug resistance and viral fitness.Antiviral Res. 2019 Nov;171:104612. doi: 10.1016/j.antiviral.2019.104612. Epub 2019 Sep 19. Antiviral Res. 2019. PMID: 31542377
-
Resistance-associated substitutions (RASs) to HCV direct-acting antivirals (DAAs) at baseline of treatment in thalassemia patients: a referral center study.Arch Virol. 2020 Oct;165(10):2193-2203. doi: 10.1007/s00705-020-04728-x. Epub 2020 Jul 8. Arch Virol. 2020. PMID: 32638116
-
Impact of Preexisting Hepatitis C Virus Genotype 6 NS3, NS5A, and NS5B Polymorphisms on the In Vitro Potency of Direct-Acting Antiviral Agents.Antimicrob Agents Chemother. 2019 Mar 27;63(4):e02205-18. doi: 10.1128/AAC.02205-18. Print 2019 Apr. Antimicrob Agents Chemother. 2019. PMID: 30718256 Free PMC article.
-
Overview of Direct-Acting Antiviral Drugs and Drug Resistance of Hepatitis C Virus.Methods Mol Biol. 2019;1911:3-32. doi: 10.1007/978-1-4939-8976-8_1. Methods Mol Biol. 2019. PMID: 30593615 Review.
-
Direct acting antivirals failure: cause and retreatment options.Expert Rev Gastroenterol Hepatol. 2018 Dec;12(12):1245-1250. doi: 10.1080/17474124.2018.1541237. Epub 2018 Oct 31. Expert Rev Gastroenterol Hepatol. 2018. PMID: 30791789 Review.
Cited by
-
Conspicuous Response to Direct-Acting Antivirals in Chronic Hepatitis C-related Immune Thrombocytopenia: A Case Report.Cureus. 2022 Apr 16;14(4):e24193. doi: 10.7759/cureus.24193. eCollection 2022 Apr. Cureus. 2022. PMID: 35592216 Free PMC article.
-
Advancing high-throughput anti-HCV drug screening: a novel dual-reporter HCV replicon model with real-time monitoring.Res Pharm Sci. 2025 Feb 20;20(1):41-54. doi: 10.4103/RPS.RPS_249_23. eCollection 2025 Feb. Res Pharm Sci. 2025. PMID: 40190821 Free PMC article.
-
Wound Healing, Antioxidant, and Antiviral Properties of Bioactive Polysaccharides of Microalgae Strains Isolated from Greek Coastal Lagoons.Mar Drugs. 2025 Feb 10;23(2):77. doi: 10.3390/md23020077. Mar Drugs. 2025. PMID: 39997201 Free PMC article.
-
Cell type dependent stability and virulence of a recombinant SARS-CoV-2, and engineering of a propagation deficient RNA replicon to analyze virus RNA synthesis.Front Cell Infect Microbiol. 2023 Oct 24;13:1268227. doi: 10.3389/fcimb.2023.1268227. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37942479 Free PMC article.
-
Generation of a SARS-CoV-2 Reverse Genetics System and Novel Human Lung Cell Lines That Exhibit High Virus-Induced Cytopathology.Viruses. 2023 May 30;15(6):1281. doi: 10.3390/v15061281. Viruses. 2023. PMID: 37376581 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

