Genomic Characterization of mcr-1- carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 Clone Isolated From Pigs in China
- PMID: 32714906
- PMCID: PMC7344297
- DOI: 10.3389/fbioe.2020.00663
Genomic Characterization of mcr-1- carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 Clone Isolated From Pigs in China
Erratum in
-
Corrigendum: Genomic Characterization of mcr-1-carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 Clone Isolated From Pigs in China.Front Bioeng Biotechnol. 2020 Aug 6;8:842. doi: 10.3389/fbioe.2020.00842. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32850716 Free PMC article.
Abstract
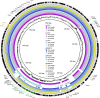
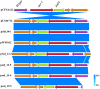
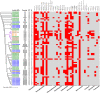
Salmonella enterica serovar 4,[5],12:i:-, so-called Typhimurium monophasic variant, has become one of the most frequently isolated serovars both in humans and in animals all over the world. The increasing prevalence of mcr-1-carrying Salmonella poses significant global health concerns. However, the potential role of Salmonella 4,[5],12:i:- in mcr-1 gene migration through the food chain to the human remains obscure. Here, we investigated 337 Salmonella isolates from apparently healthy finishing pigs, which is rarely studied, obtained from pig farms and slaughterhouses in China. The mcr-1 gene was found in four colistin-resistant S. enterica 4,[5],12:i:- isolates. Notably, all four isolates belonged to sequence type 34 (ST34) with multidrug resistance phenotype. Further genomic sequencing and antimicrobial resistance characterization confirmed that mcr was responsible for the colistin resistance, and the conjugation assay demonstrated that three of four isolates carried mcr-1 in IncHI2 plasmid. Importantly, mcr-1 and class-1 integron were found to co-localize in two strains with IncHI2 plasmid. By collecting all the mcr-1-carrying Typhimurium and monophasic variant strains across the food chain (farm animals, animal-origin food, and humans), our phylogenomic analysis of available 66 genomes, including four strains in this study, demonstrated an independent phylogenetic cluster of all eight Chinese swine-originated isolates and one human isolate. Together, this study provides direct evidence for clonal and pork-borne transmission of mcr-1 by Salmonella 4,[5],12:i:- ST34 in China and highlighted a domestication pathway by acquisition of additional antimicrobial resistance determinants in Chinese ST34 isolates.
Keywords: 12:i:-; Colistin; ST34; Salmonella enterica serovar 4; [5]; food chain; mcr-1.
Copyright © 2020 Elbediwi, Pan, Jiang, Biswas, Li and Yue.
Figures

References
LinkOut - more resources
Full Text Sources

