Congruence of Transcription Programs in Adult Stem Cell-Derived Jejunum Organoids and Original Tissue During Long-Term Culture
- PMID: 32714922
- PMCID: PMC7343960
- DOI: 10.3389/fcell.2020.00375
Congruence of Transcription Programs in Adult Stem Cell-Derived Jejunum Organoids and Original Tissue During Long-Term Culture
Abstract
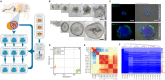
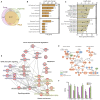
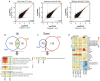
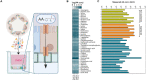
The emergence of intestinal organoids, as a stem cell-based self-renewable model system, has led to many studies on intestinal development and cell-cell signaling. However, potential issues regarding the phenotypic stability and reproducibility of the methodology during culture still needs to be addressed for different organoids. Here we investigated the transcriptomes of jejunum organoids derived from the same pig as well as batch-to-batch variation of organoids derived from different pigs over long-term passage. The set of genes expressed in organoids closely resembled that of the tissue of origin, including small intestine specific genes, for at least 17 passages. Minor differences in gene expression were observed between individual organoid cultures. In contrast, most small intestine-specific genes were not expressed in the jejunum cell line IPEC-J2, which also showed gene expression consistent with cancer phenotypes. We conclude that intestinal organoids provide a robust and stable model for translational research with clear advantages over transformed cells.
Keywords: IPEC-J2; gastrointestinal; intestinal organoids; organoid stability; porcine organoids.
Copyright © 2020 van der Hee, Madsen, Vervoort, Smidt and Wells.
Figures


References
-
- Andrews S. (2017). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

