The evolution and functional divergence of the histone H2B family in plants
- PMID: 32716939
- PMCID: PMC7410336
- DOI: 10.1371/journal.pgen.1008964
The evolution and functional divergence of the histone H2B family in plants
Abstract
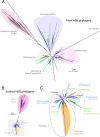
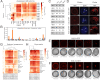
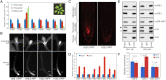
Chromatin regulation of eukaryotic genomes depends on the formation of nucleosome complexes between histone proteins and DNA. Histone variants, which are diversified by sequence or expression pattern, can profoundly alter chromatin properties. While variants in histone H2A and H3 families are well characterized, the extent of diversification of histone H2B proteins is less understood. Here, we report a systematic analysis of the histone H2B family in plants, which have undergone substantial divergence during the evolution of each major group in the plant kingdom. By characterising Arabidopsis H2Bs, we substantiate this diversification and reveal potential functional specialization that parallels the phylogenetic structure of emergent clades in eudicots. In addition, we identify a new class of highly divergent H2B variants, H2B.S, that specifically accumulate during chromatin compaction of dry seed embryos in multiple species of flowering plants. Our findings thus identify unsuspected diverse properties among histone H2B proteins in plants that has manifested into potentially novel groups of histone variants.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

