Gene expression profiling of hypertrophic cardiomyocytes identifies new players in pathological remodelling
- PMID: 32717063
- PMCID: PMC8152696
- DOI: 10.1093/cvr/cvaa233
Gene expression profiling of hypertrophic cardiomyocytes identifies new players in pathological remodelling
Abstract
Aims: Pathological cardiac remodelling is characterized by cardiomyocyte (CM) hypertrophy and fibroblast activation, which can ultimately lead to maladaptive hypertrophy and heart failure (HF). Genome-wide expression analysis on heart tissue has been instrumental for the identification of molecular mechanisms at play. However, these data were based on signals derived from all cardiac cell types. Here, we aimed for a more detailed view on molecular changes driving maladaptive CM hypertrophy to aid in the development of therapies to reverse pathological remodelling.
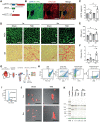
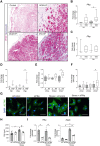
Methods and results: Utilizing CM-specific reporter mice exposed to pressure overload by transverse aortic banding and CM isolation by flow cytometry, we obtained gene expression profiles of hypertrophic CMs in the more immediate phase after stress, and CMs showing pathological hypertrophy. We identified subsets of genes differentially regulated and specific for either stage. Among the genes specifically up-regulated in the CMs during the maladaptive phase we found known stress markers, such as Nppb and Myh7, but additionally identified a set of genes with unknown roles in pathological hypertrophy, including the platelet isoform of phosphofructokinase (PFKP). Norepinephrine-angiotensin II treatment of cultured human CMs induced the secretion of N-terminal-pro-B-type natriuretic peptide (NT-pro-BNP) and recapitulated the up-regulation of these genes, indicating conservation of the up-regulation in failing CMs. Moreover, several genes induced during pathological hypertrophy were also found to be increased in human HF, with their expression positively correlating to the known stress markers NPPB and MYH7. Mechanistically, suppression of Pfkp in primary CMs attenuated stress-induced gene expression and hypertrophy, indicating that Pfkp is an important novel player in pathological remodelling of CMs.
Conclusion: Using CM-specific transcriptomic analysis, we identified novel genes induced during pathological hypertrophy that are relevant for human HF, and we show that PFKP is a conserved failure-induced gene that can modulate the CM stress response.
Keywords: PFKP; Cardiomyocyte; Heart failure; Hypertrophy; Pathological remodelling; RNA sequencing.
© The Author(s) 2020. Published by Oxford University Press on behalf of the European Society of Cardiology.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

