Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology
- PMID: 32717794
- PMCID: PMC7463467
- DOI: 10.3390/polym12081633
Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology
Abstract
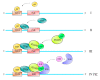
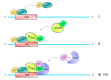
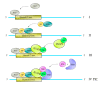
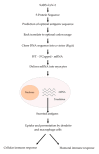
Multi-subunit enzymes are protein biopolymers that are involved in many cellular processes. The enzyme that carries out the process of transcription of mRNAs is RNA polymerase II (RNAPII), which is a multi-subunit enzyme in eukaryotes. This protein biopolymer starts the transcription from specific sites and is positioned by transcription factors, which form a preinitiation complex (PIC) on gene promoters. To recognize and position the RNAPII and the transcription factors on the gene promoters are needed specific DNA sequences in the gene promoters, which are named promoter elements. Those gene promoter elements can vary and therefore several kinds of promoters exist, however, it appears that all promoters can use a similar pathway for PIC formation. Those pathways are discussed in this review. The in vitro transcribed mRNA can be used as vaccines to fight infectious diseases, e.g., in immunotherapy against cancer and in nanotechnology to deliver mRNA for a missing protein into the cell. We have outlined a procedure to produce an mRNA vaccine against the SARS-CoV-2 virus, which is the causing agent of the big pandemic, COVID-19, affecting human beings all over the world. The potential advantages of using eukaryotic RNAPII to synthetize large transcripts are outlined and discussed. In addition, we suggest a method to cap the mRNA at the 5' terminus by using enzymes, which might be more effective than cap analogs. Finally, we suggest the construction of a future multi-talented RNAPII, which would be able to synthetize large mRNA and cap them in the test tube.
Keywords: immunotherapy; mRNA; nanotechnology; protein biopolymer; transcription; vaccine.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

