Bayesian model selection reveals biological origins of zero inflation in single-cell transcriptomics
- PMID: 32718323
- PMCID: PMC7384222
- DOI: 10.1186/s13059-020-02103-2
Bayesian model selection reveals biological origins of zero inflation in single-cell transcriptomics
Erratum in
-
Publisher Correction: Bayesian model selection reveals biological origins of zero inflation in single-cell transcriptomics.Genome Biol. 2020 Nov 3;21(1):270. doi: 10.1186/s13059-020-02182-1. Genome Biol. 2020. PMID: 33143736 Free PMC article.
Abstract
Background: Single-cell RNA sequencing is a powerful tool for characterizing cellular heterogeneity in gene expression. However, high variability and a large number of zero counts present challenges for analysis and interpretation. There is substantial controversy over the origins and proper treatment of zeros and no consensus on whether zero-inflated count distributions are necessary or even useful. While some studies assume the existence of zero inflation due to technical artifacts and attempt to impute the missing information, other recent studies argue that there is no zero inflation in scRNA-seq data.
Results: We apply a Bayesian model selection approach to unambiguously demonstrate zero inflation in multiple biologically realistic scRNA-seq datasets. We show that the primary causes of zero inflation are not technical but rather biological in nature. We also demonstrate that parameter estimates from the zero-inflated negative binomial distribution are an unreliable indicator of zero inflation.
Conclusions: Despite the existence of zero inflation in scRNA-seq counts, we recommend the generalized linear model with negative binomial count distribution, not zero-inflated, as a suitable reference model for scRNA-seq analysis.
Keywords: Bayesian model selection; Cell heterogeneity; Gene expression stochasticity; Single-cell RNA sequencing; Zero inflation.
Conflict of interest statement
None of the authors declares any competing interests.
Figures
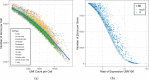

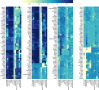
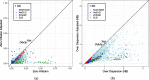

References
-
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11(2):163–6. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

