Functional annotation of human long noncoding RNAs via molecular phenotyping
- PMID: 32718982
- PMCID: PMC7397864
- DOI: 10.1101/gr.254219.119
Functional annotation of human long noncoding RNAs via molecular phenotyping
Abstract
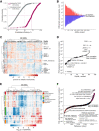
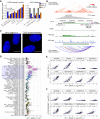
Long noncoding RNAs (lncRNAs) constitute the majority of transcripts in the mammalian genomes, and yet, their functions remain largely unknown. As part of the FANTOM6 project, we systematically knocked down the expression of 285 lncRNAs in human dermal fibroblasts and quantified cellular growth, morphological changes, and transcriptomic responses using Capped Analysis of Gene Expression (CAGE). Antisense oligonucleotides targeting the same lncRNAs exhibited global concordance, and the molecular phenotype, measured by CAGE, recapitulated the observed cellular phenotypes while providing additional insights on the affected genes and pathways. Here, we disseminate the largest-to-date lncRNA knockdown data set with molecular phenotyping (over 1000 CAGE deep-sequencing libraries) for further exploration and highlight functional roles for ZNF213-AS1 and lnc-KHDC3L-2.
© 2020 Ramilowski et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
