Quantitating denaturation by formic acid: imperfect repeats are essential to the stability of the functional amyloid protein FapC
- PMID: 32719003
- PMCID: PMC7489911
- DOI: 10.1074/jbc.RA120.013396
Quantitating denaturation by formic acid: imperfect repeats are essential to the stability of the functional amyloid protein FapC
Abstract
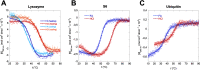
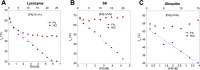
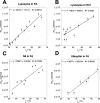
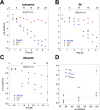
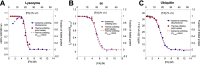
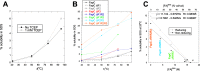
Bacterial functional amyloids are evolutionarily optimized to aggregate, so much so that the extreme robustness of functional amyloid makes it very difficult to examine their structure-function relationships in a detailed manner. Previous work has shown that functional amyloids are resistant to conventional chemical denaturants, but they dissolve in formic acid (FA) at high concentrations. However, systematic investigation requires a quantitative analysis of FA's ability to denature proteins. Amyloid formed by Pseudomonas sp. protein FapC provides an excellent model to investigate FA denaturation. It contains three imperfect repeats, and stepwise removal of these repeats slows fibrillation and increases fragmentation during aggregation. However, the link to stability is unclear. We first calibrated FA denaturation using three small, globular, and acid-resistant proteins. This revealed a linear relationship between the concentration of FA and the free energy of unfolding with a slope of mFA+pH (the combined contribution of FA and FA-induced lowering of pH), as well as a robust correlation between protein size and mFA+pH We then measured the solubilization of fibrils formed from different FapC variants with varying numbers of repeats as a function of the concentration of FA. This revealed a decline in the number of residues driving amyloid formation upon deleting at least two repeats. The midpoint of denaturation declined with the removal of repeats. Complete removal of all repeats led to fibrils that were solubilized at FA concentrations 2-3 orders of magnitude lower than the repeat-containing variants, showing that at least one repeat is required for the stability of functional amyloid.
Keywords: FapC; Pseudomonas; fibril; formic acid; functional amyloid; heat capacity; m-values; protein denaturation; protein stability; thermodynamics.
© 2020 Christensen et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Dueholm M., Nielsen P. H., Chapman M. R., and Otzen D. E. (2012) Functional amyloids in Bacteria. in Amyloid Fibrils and Prefibrillar Aggregates (Otzen D. E., ed.), pp. 411–438, Wiley-VCH Verlag GmbH, Berlin, Germany
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

