3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing
- PMID: 32719531
- PMCID: PMC7537785
- DOI: 10.1038/s41592-020-0890-0
3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing
Abstract
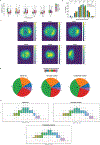
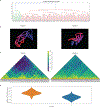
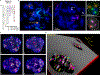
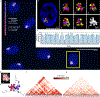
There is a need for methods that can image chromosomes with genome-wide coverage, as well as greater genomic and optical resolution. We introduce OligoFISSEQ, a suite of three methods that leverage fluorescence in situ sequencing (FISSEQ) of barcoded Oligopaint probes to enable the rapid visualization of many targeted genomic regions. Applying OligoFISSEQ to human diploid fibroblast cells, we show how four rounds of sequencing are sufficient to produce 3D maps of 36 genomic targets across six chromosomes in hundreds to thousands of cells, implying a potential to image thousands of targets in only five to eight rounds of sequencing. We also use OligoFISSEQ to trace chromosomes at finer resolution, following the path of the X chromosome through 46 regions, with separate studies showing compatibility of OligoFISSEQ with immunocytochemistry. Finally, we combined OligoFISSEQ with OligoSTORM, laying the foundation for accelerated single-molecule super-resolution imaging of large swaths of, if not entire, human genomes.
Conflict of interest statement
Competing Interests:
Harvard University has filed patent applications on behalf of C-tW, HQN, and SC, pertaining to Oligopaints and related oligo-based methods for genome imaging. ERD is currently an employee of ReadCoor and has an equity interest in ReadCoor. Potential conflicts of interests for GMC are listed on
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

